| Original Literature | Model OverView |
|---|---|
|
Publication
Title
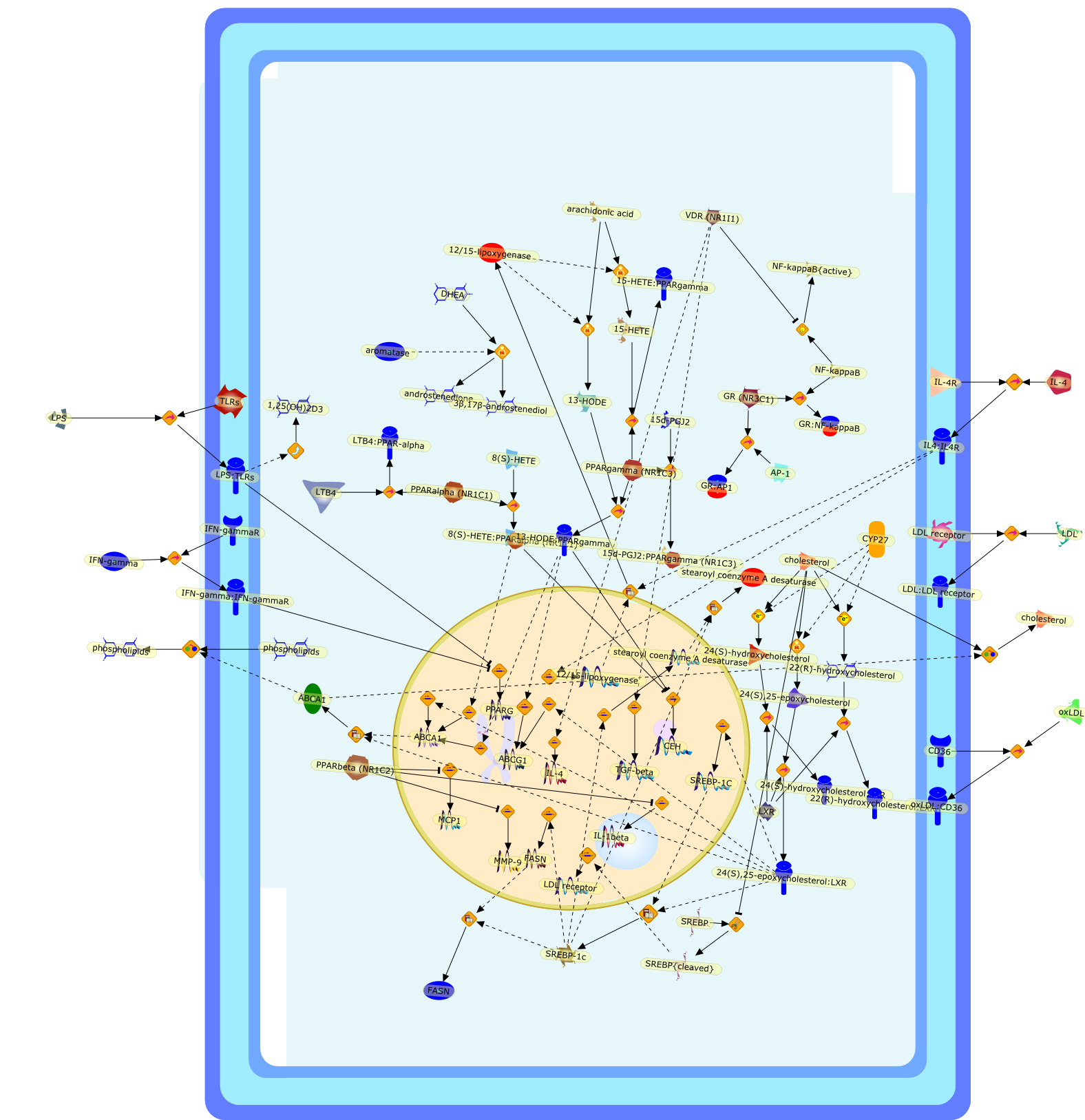
Nuclear receptor signaling in macrophages.
Affiliation
Department of Cellular and Molecular Medicine, University of California, SanDiego, 9500 Gilman Drive, La Jolla, CA 92093-0651, USA.
Abstract
Macrophages play diverse roles in host defense and in maintenance ofhomeostasis. Based on their ability to promote inflammatory responses,inappropriate macrophage function also contributes to numerous pathologicalprocesses, including atherosclerosis, rheumatoid arthritis and inflammatorybowel disease. Members of the nuclear receptor superfamily of ligand-dependenttranscriptions factors have emerged as key regulators of inflammation and lipidhomeostasis in macrophages. These include the glucocorticoid receptor (GR),which inhibits inflammatory programs of gene expression in response to naturalcorticosteroids and synthetic anti-inflammatory ligands such as dexamethasone.Also, in response to endogenous eicosanoids and oxysterols, respectively,peroxisome proliferator-activated receptors (PPARs) and liver X receptors (LXRs)regulate transcriptional programs involved in inflammatory responses and lipidhomeostasis. Identification of their mechanisms of action should help guide thedevelopment of new therapeutic agents useful in the treatment of diseases inwhich macrophages play critical pathogenic roles.
PMID
14698033
|