| Original Literature | Model OverView |
|---|---|
|
Publication
Title
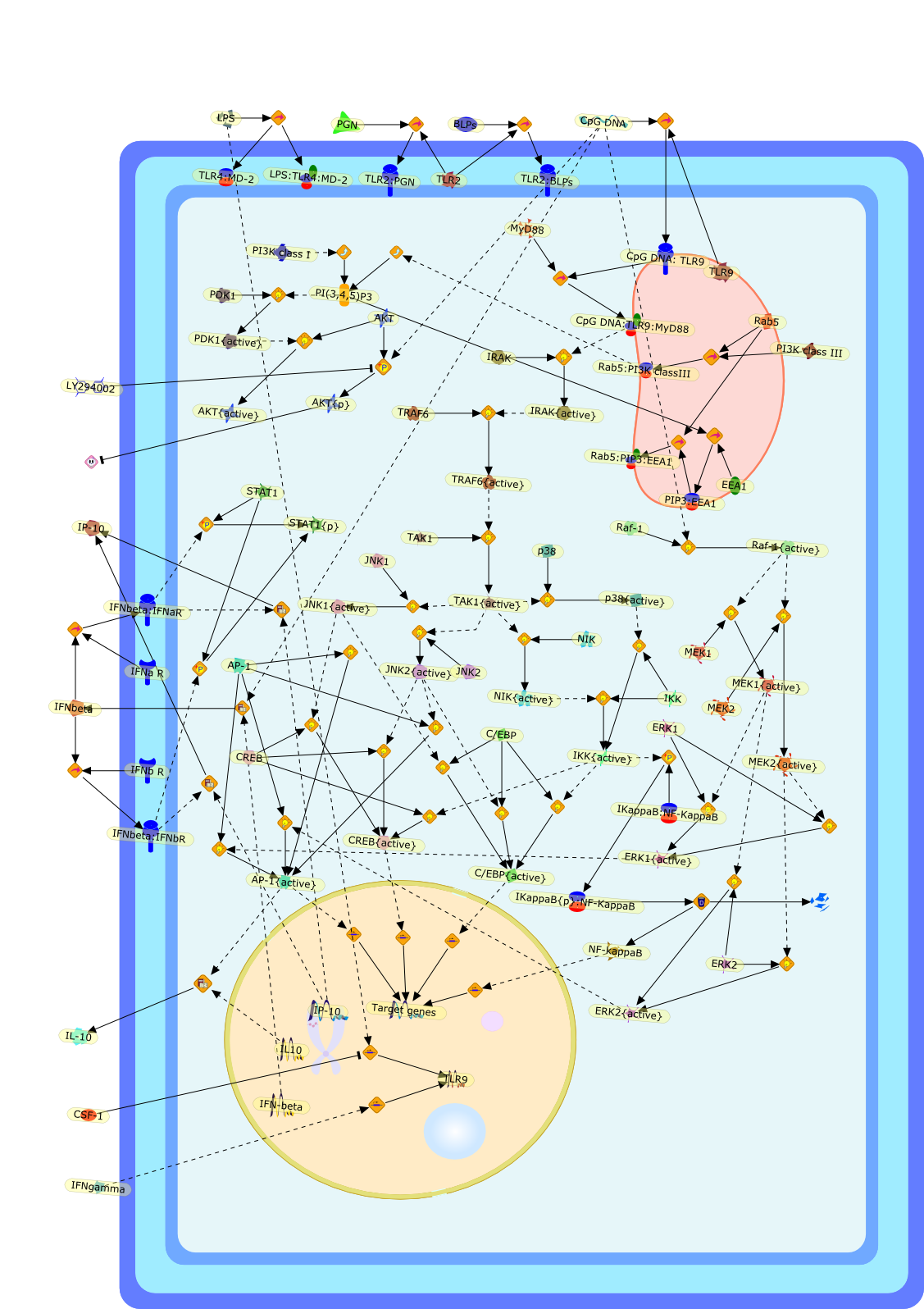
Signal transduction pathways mediated by the interaction of CpG DNA withToll-like receptor 9.
Affiliation
Center for Biologics Evaluation and Research/Food and Drug Administration, Bldg29A, Rm 3D10, Bethesda, MD 20892, USA.
Abstract
Synthetic oligodeoxynucleotides (ODN) expressing non-methylated "CpG motifs"patterned after those present in bacterial DNA have characteristicimmunomodulatory effects. CpG DNA is recognized as a pathogen-associatedmolecular pattern, and triggers a rapid innate immune response. CpG ODN arebeing harnessed for a variety of therapeutic uses, including as immuneadjuvants, for cancer therapy, as anti-allergens, and as immunoprotectiveagents. The signal transduction pathway mediated by the engagement of CpG DNAwith Toll-like receptor 9 (TLR9) is shared with other members of the TLR family.Recent studies demonstrate that formation and maturation of CpG DNA-containingendosomes are regulated by phosphatidylinositol 3 kinases and the Ras-associatedGTP-binding protein, Rab5, which are essential for the initiation ofTLR9-mediated signaling.
PMID
14751759
|