| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Innate immunity and toll-like receptors: clinical implications of basic scienceresearch.
Affiliation
Inflammatory Bowel Disease Center, Division of Gastroenterology, Department ofMedicine, Steven Spielberg Pediatric Research Center, Los Angeles, CA 90048,USA.
Abstract
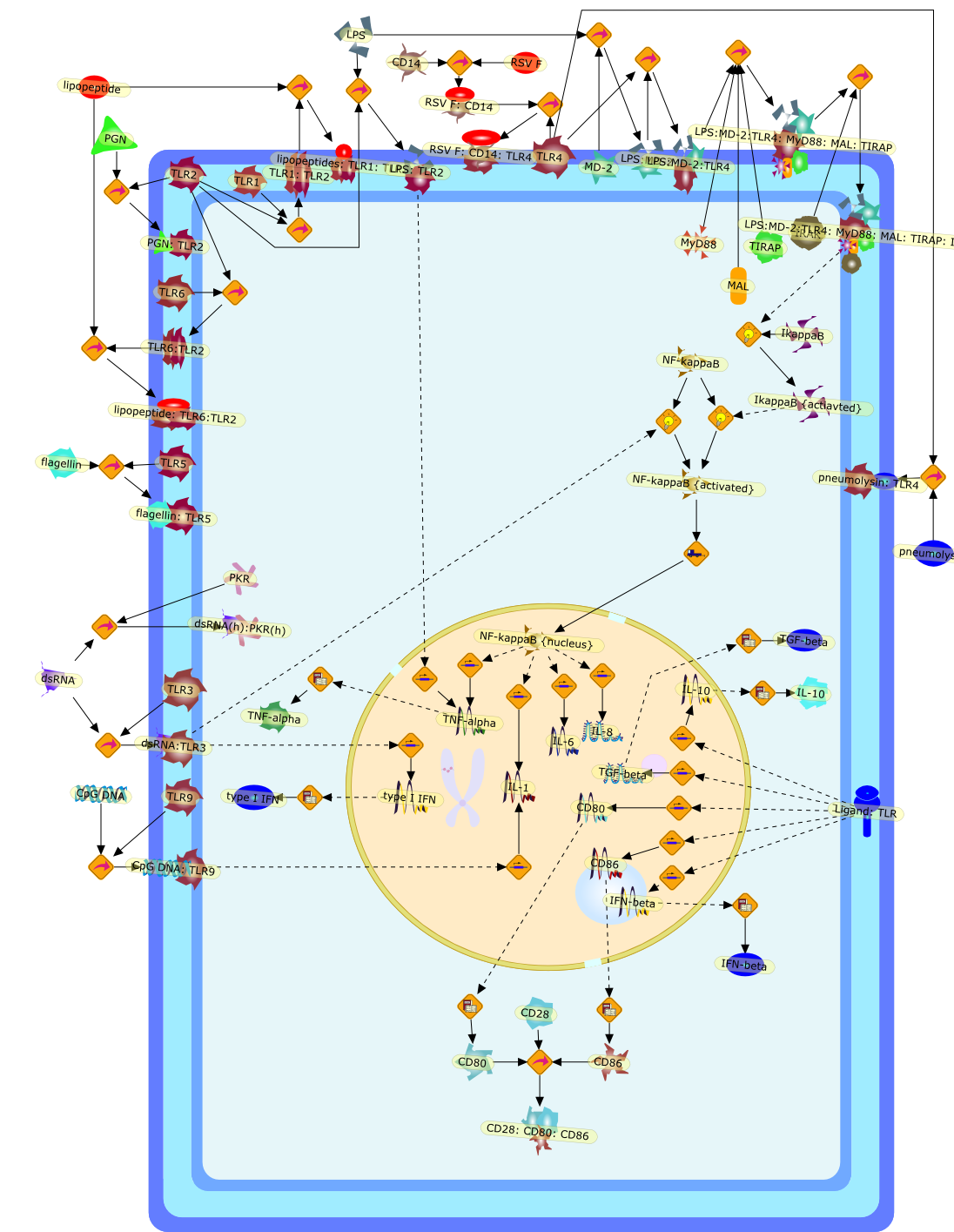
Humans are constantly exposed to a wide variety of microorganisms that can causeinfection. In self-defense, the human host has evolved complex protectivemechanisms, and Toll-like receptors (TLRs) have emerged as a central point indefense. These receptors bind molecular structures that are expressed bymicrobes but are not expressed by the human host, eg, lipopolysaccharides (LPS)or double-stranded RNA (dsRNA). Activation of these receptors initiates aninflammatory cascade that attempts to clear the offending pathogen and set inmotion a specific adaptive immune response. Defects in sensing of pathogens maypredispose the host to recurrent infections. The relative rarity of thesesyndromes of defective innate immunity, however, speaks to the redundancy insensing of pathogens by the innate immune system. More common, polymorphisms inTLR4 are associated with increased predisposition to severe and recurrentinfections but protection against atherosclerotic disease due to diminishedinflammation. Toll-like receptor signaling may also contribute to thepathophysiology of disease and injure the host by activating a deleteriousimmune response such as in sepsis or inflammatory bowel disease (IBD). The focusof this article is to describe the role of TLRs in the innate immune response inhealth and disease.
PMID
15069387
|