| Original Literature | Model OverView |
|---|---|
|
Publication
Title
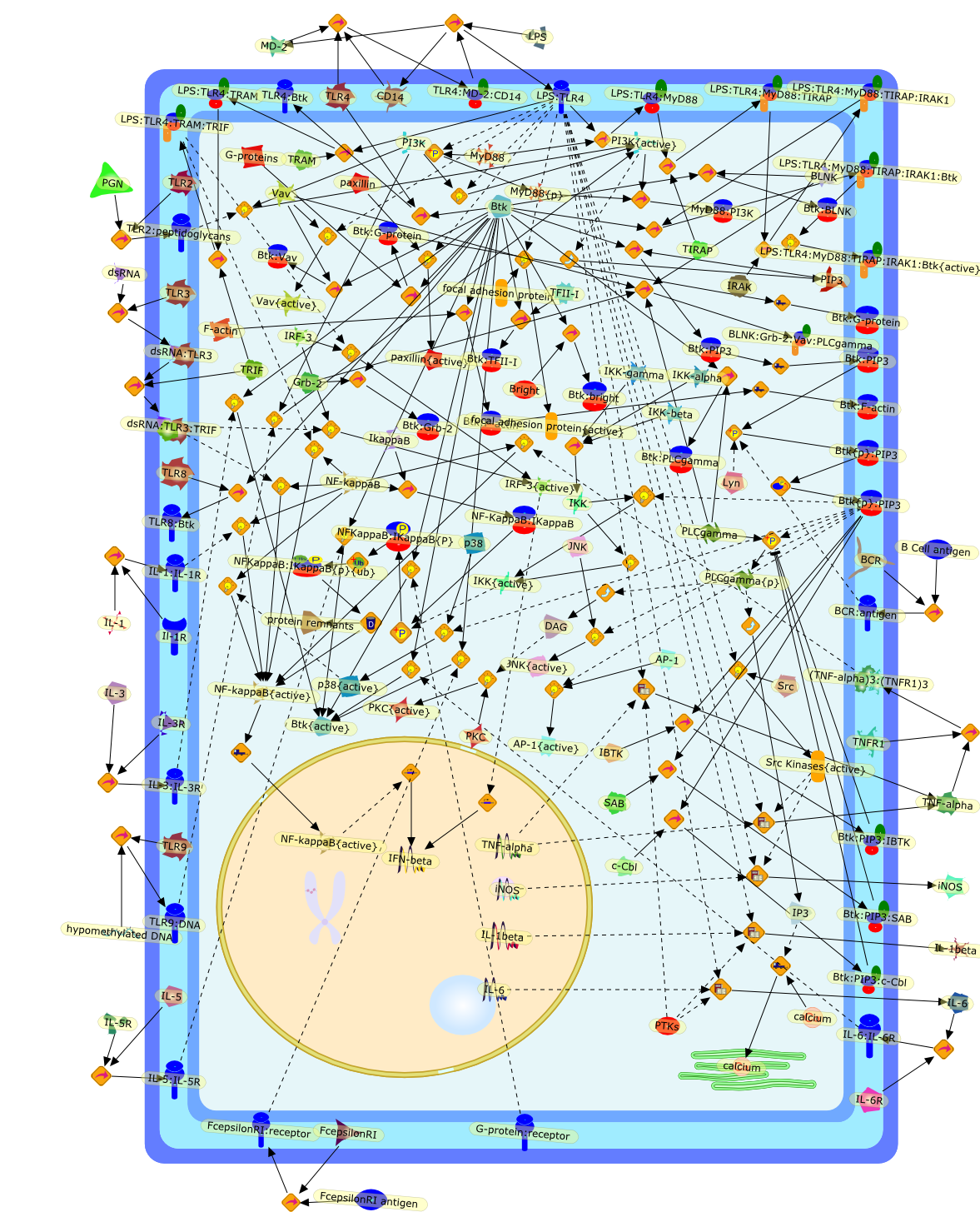
Bruton's tyrosine kinase (Btk)-the critical tyrosine kinase in LPS signalling?
Affiliation
Department of Biochemistry and Biotechnology Institute, Cytokine Research Group,Trinity College, Dublin 2, Ireland. jefferca@tcd.ie
Abstract
The discovery of the Toll-like receptors (TLRs) has revolutionised the field ofinnate immunity. One unresolved question regarding LPS signalling is whetherthere is a role for tyrosine kinases downstream of the LPS receptor. Studies inmice deficient in Bruton's tyrosine kinase have previously shown that they aredefective in their responses to LPS. Further investigation into the role of Btkin LPS signalling has directly implicated Btk downstream of TLR4, both withrespect to p38 MAPK activation and activation of the transcription factorNFkappaB. In fact Btk is activated by LPS and has been shown to directly bindTLR4 and the key proximal signalling proteins involved in LPS-induced NFkappaBactivation, MyD88, Mal and IRAK-1. These recent findings point to a direct rolefor Btk in LPS signal transduction and raise interesting questions regarding themode of activation of Btk following LPS stimulation and the precise nature ofthe pathways activated downstream of Btk. A better understanding of how Btkfunctions in LPS signalling will have important implications for inflammatoryand autoimmune disorders and therapies thereof.
PMID
15081522
|