| Original Literature | Model OverView |
|---|---|
|
Publication
Title
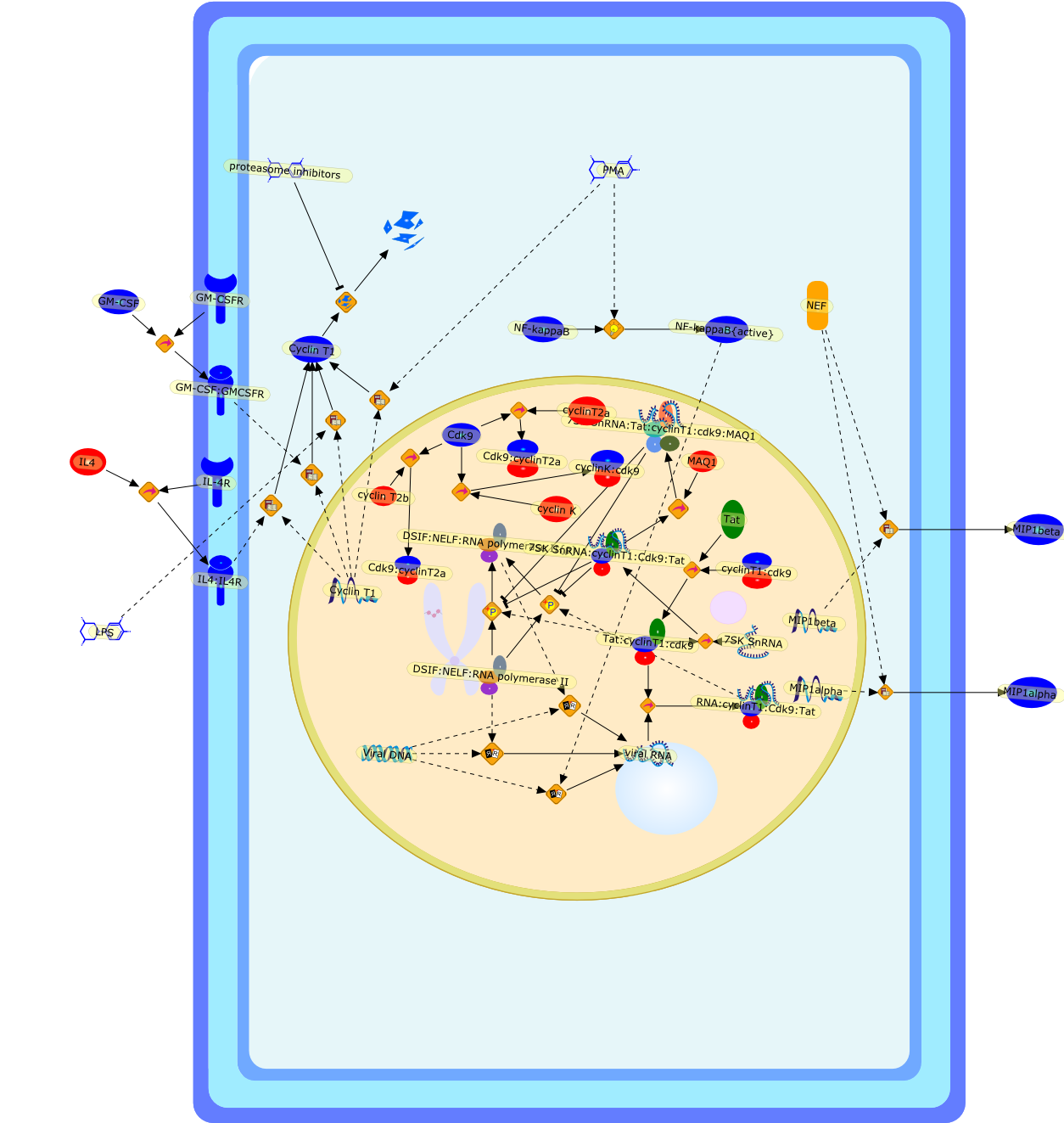
HIV-1 infection and regulation of Tat function in macrophages.
Affiliation
Department of Molecular Virology and Microbiology, Baylor College of Medicine,One Baylor Plaza, Houston, TX 77030, USA.
Abstract
The macrophage is an important cell type in the pathophysiology of humanimmunodeficiency virus type 1 (HIV-1) infection. Macrophages both support viralreplication and are capable of attracting and activating lymphocytes, thusrendering CD4+ T lymphocytes highly permissive for infection. The viral Tatprotein, whose function is mediated by the cellular cyclin T1 protein complexedwith CDK9, is required for efficient transcription of the integrated HIV-1provirus by RNA polymerase II. Cyclin T1 expression is highly regulated duringmacrophage differentiation, and this has important implications for HIV-1replication. In monocytes isolated from healthy blood donors, cyclin T1 proteinexpression is low and is induced to high levels within the first few days ofdifferentiation by a post-transcriptional mechanism. After 1-2 weeks ofmacrophage differentiation, however, cyclin T1 expression is shut off. Treatmentof macrophages with lipopolysaccharide (LPS) can re-induce cyclin T1, indicatingthat the activation status of macrophages can regulate cyclin T1 expression.Recent results indicate that HIV-1 infection is able to induce cyclin T1expression in macrophages. Future studies of cyclin T1 regulation in macrophagesmay suggest means of manipulating expression of this crucial cellular co-factorfor therapeutic benefit in HIV-1 infected individuals. Copyright 2004 ElsevierB.V.
PMID
15183343
|