| Original Literature | Model OverView |
|---|---|
|
Publication
Title
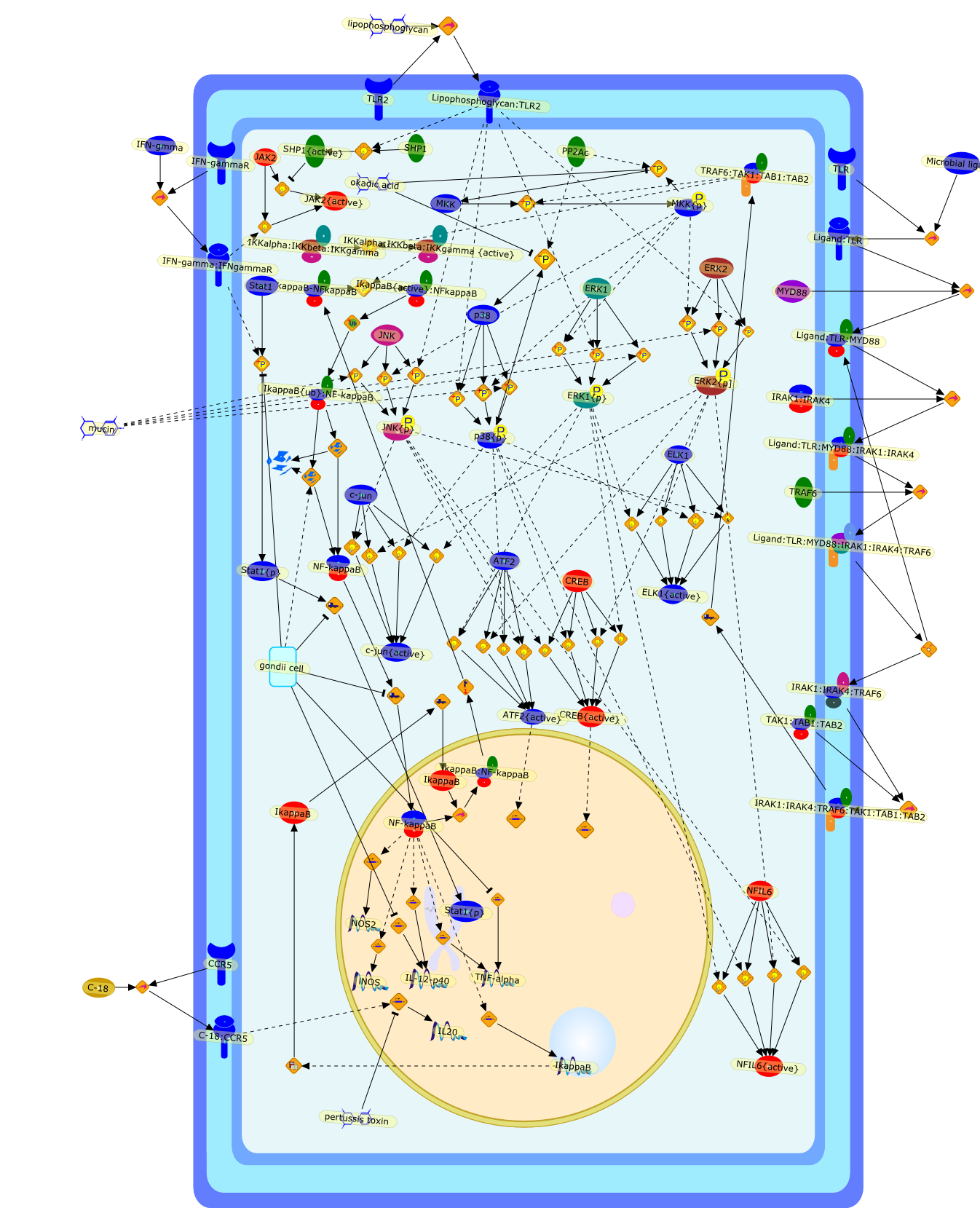
Manipulation of mitogen-activated protein kinase/nuclear factor-kappaB-signalingcascades during intracellular Toxoplasma gondii infection.
Affiliation
Department of Microbiology and Immunology, College of Veterinary Medicine,Cornell University, Ithaca, NY, USA. eyd1@cornell.edu
Abstract
The intracellular protozoan Toxoplasma gondii exerts profound effects on nuclearfactor-kappaB (NF-kappaB)- and mitogen-activated protein kinase (MAPK)-signalingcascades in macrophages. During early infection, nuclear translocation ofNF-kappaB is blocked, and later, the cells display defects in lipopolysaccharide(LPS)-induced MAPK phosphorylation after undergoing initial activation inresponse to Toxoplasma itself. Infected macrophages that are subjected totriggering through Toll-like receptor 4 (TLR4) with LPS display defectiveproduction of tumor necrosis factor-alpha and IL-12 (IL-12) that likely reflectsinterference with NF-kappaB- and MAPK-signaling cascades. Nevertheless, T.gondii possesses molecules that themselves induce eventual proinflammatorycytokine synthesis. For interleukin-12, this occurs through both myeloiddifferentiation factor 88-dependent and chemokine receptor CCR5-dependentpathways. The balance between activation and interference with proinflammatorysignaling is likely to reflect the need to achieve an appropriate level ofimmunity that allows the host and parasite to maintain a stable interaction.
PMID
15361242
|