| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Inhibition of toll-like receptor and cytokine signaling--a unifying theme inischemic tolerance.
Affiliation
Department of Neurosurgery, University of Pennsylvania School of Medicine,Philadelphia, Pennsylvania, USA. kariko@mail.med.upenn.edu
Abstract
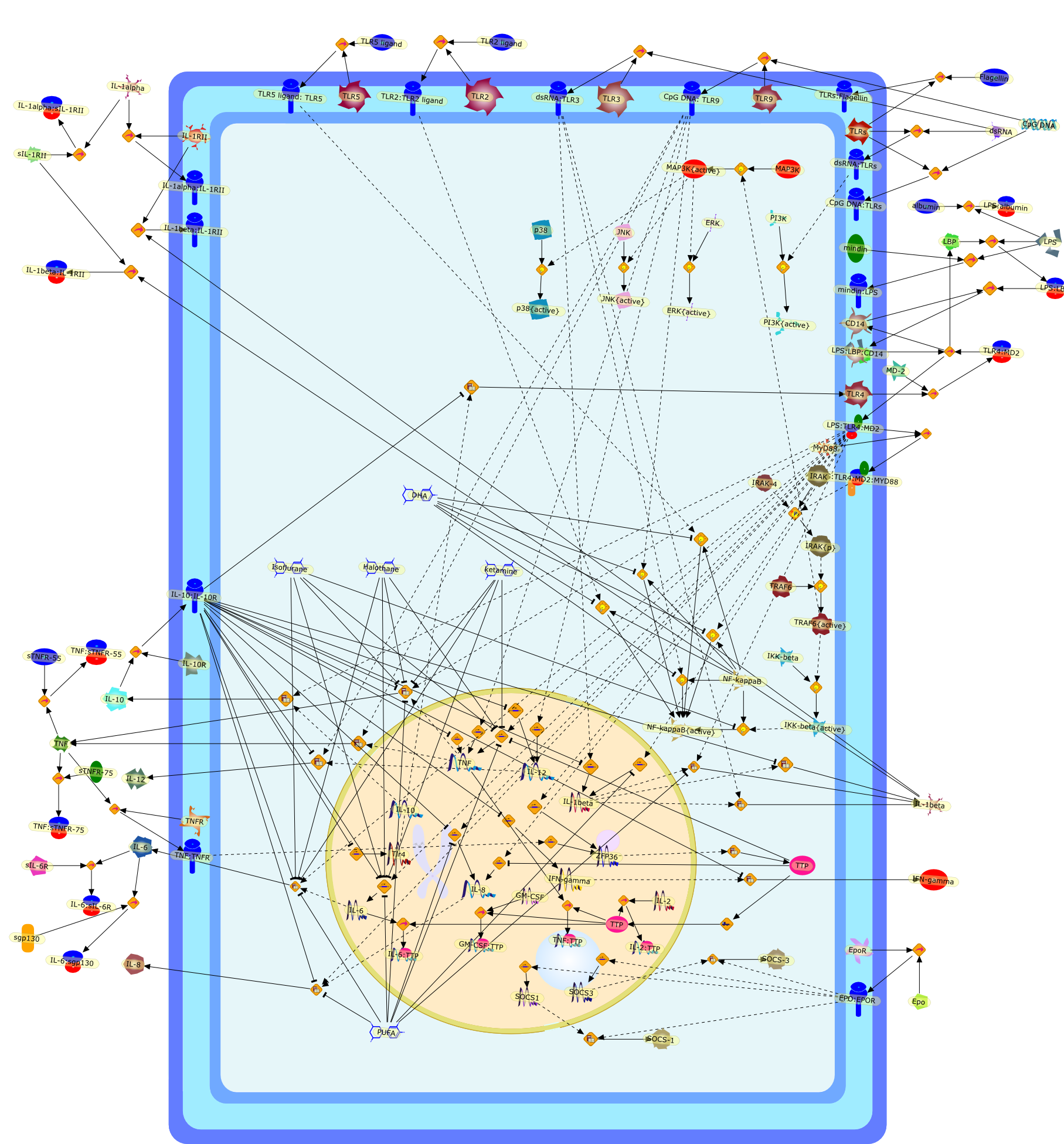
Cerebral ischemia triggers acute inflammation, which exacerbates primary braindamage. Activation of the innate immune system is an important component of thisinflammatory response. Inflammation occurs through the action of proinflammatorycytokines, such as TNF, IL-1 beta and IL-6, that alter blood flow and increasevascular permeability, thus leading to secondary ischemia and accumulation ofimmune cells in the brain. Production of these cytokines is initiated bysignaling through Toll-like receptors (TLRs) that recognize host-derivedmolecules released from injured tissues and cells. Recently, great strides havebeen made in understanding the regulation of the innate immune system,particularly the signaling mechanisms of TLRs. Negative feedback inhibitors ofTLRs and inflammatory cytokines have now been identified and characterized. Itis also evident that lipid rafts exist in membranes and play a role inreceptor-mediated inflammatory signaling events. In the present review, usingthis newly available large body of knowledge, we take a fresh look at studies ofischemic tolerance. Based on this analysis, we recognize a striking similaritybetween ischemic tolerance and endotoxin tolerance, an immune suppressive statecharacterized by hyporesponsiveness to lipopolysaccharide (LPS). In view of thisanalogy, and considering recent discoveries related to molecular mechanisms ofendotoxin tolerance, we postulate that inhibition of TLR and proinflammatorycytokine signaling contributes critically to ischemic tolerance in the brain andother organs. Ischemic tolerance is a protective mechanism induced by a varietyof preconditioning stimuli. Tolerance can be established with two temporalprofiles: (i) a rapid form in which the trigger induces tolerance to ischemiawithin minutes and (ii) a delayed form in which development of protection takesseveral hours or days and requires de-novo protein synthesis. The rapid form oftolerance is achieved by direct interference with membrane fluidity, causingdisruption of lipid rafts leading to inhibition of TLR/cytokine signalingpathways. In the delayed form of tolerance, the preconditioning stimulus firsttriggers the TLR/cytokine inflammatory pathways, leading not only toinflammation but also to simultaneous upregulation of feedback inhibitors ofinflammation. These inhibitors, which include signaling inhibitors, decoyreceptors, and anti-inflammatory cytokines, reduce the inflammatory response toa subsequent episode of ischemia. This novel interpretation of the molecularmechanism of ischemic tolerance highlights new avenues for future investigationinto the prevention and treatment of stroke and related diseases.
PMID
15545925
|