| Original Literature | Model OverView |
|---|---|
|
Publication
Title
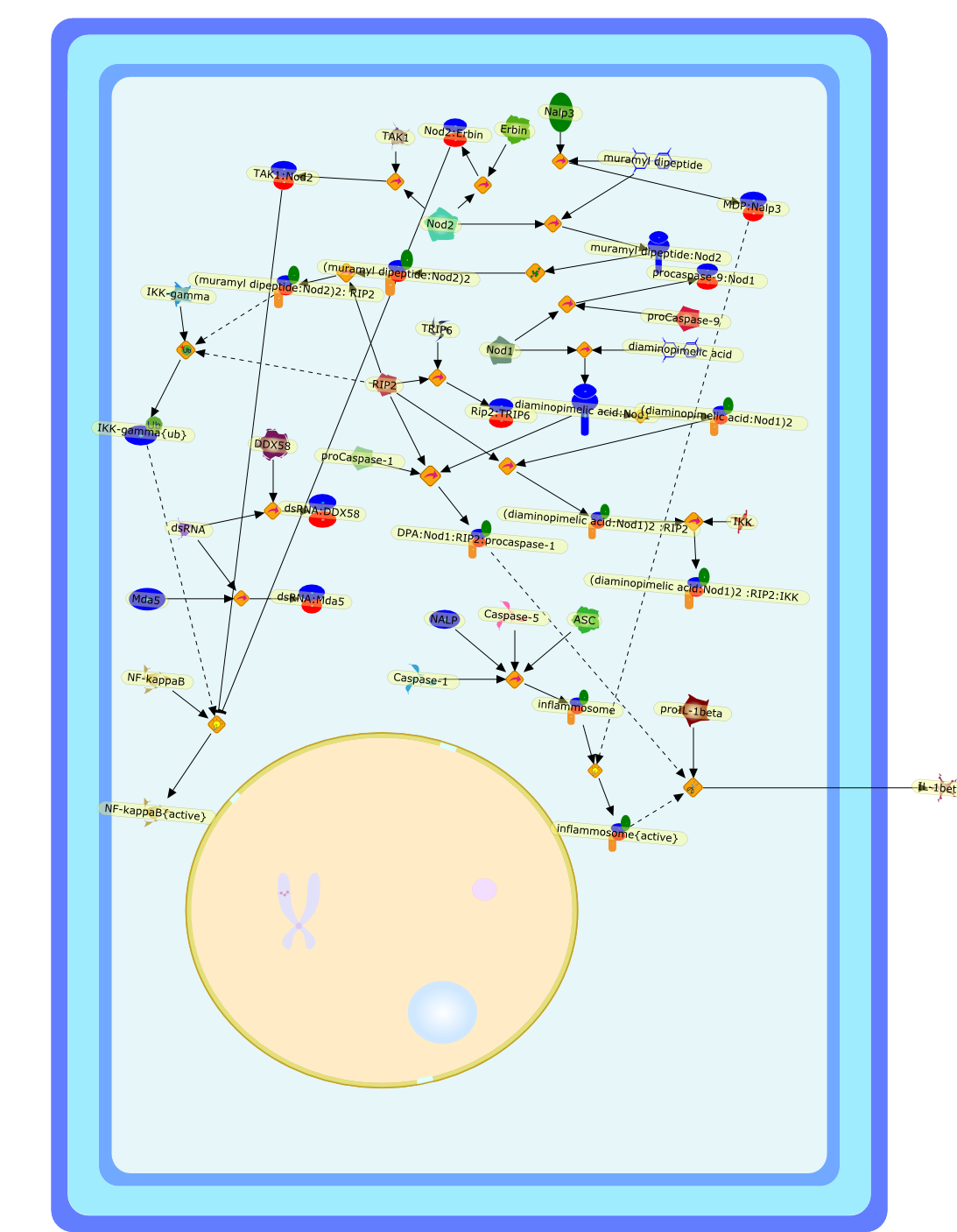
Role of Nods in bacterial infection.
Affiliation
Innate Immunity and Signalisation, Institut Pasteur, 28, Rue du Dr. Roux, 75724Paris Cedex 15, France.
Abstract
Research into intracellular sensing of microbial products is an up and comingfield in innate immunity. Nod1 and Nod2 are members of the rapidly expandingfamily of NACHT domain-containing proteins involved in intracellular recognitionof bacterial products. Nods proteins are involved in the cytosolic detection ofpeptidoglycan motifs of bacteria, recognized through the LRR domain. The role ofthe NACHT-LRR system of detection in innate immune responses is highlighted atthe mucosal barrier, where most of the membranous Toll like receptors (TLRs) arenot expressed, or with pathogens that have devised ways to escape TLR sensing.For a given pathogen, the sum of the pathways induced by the recognition of thedifferent "pathogen associated molecular patterns" (PAMPs) by the differentpattern recognition receptors (PRRs) trigger and shape the subsequent innate andadaptive immune responses. Knowledge gathered during the last decade on PRR andtheir agonists, and recent studies on bacterial infections provide new insightsinto the immune response and the pathogenesis of human infectious diseases.
PMID
17379560
|