| Original Literature | Model OverView |
|---|---|
|
Publication
Title
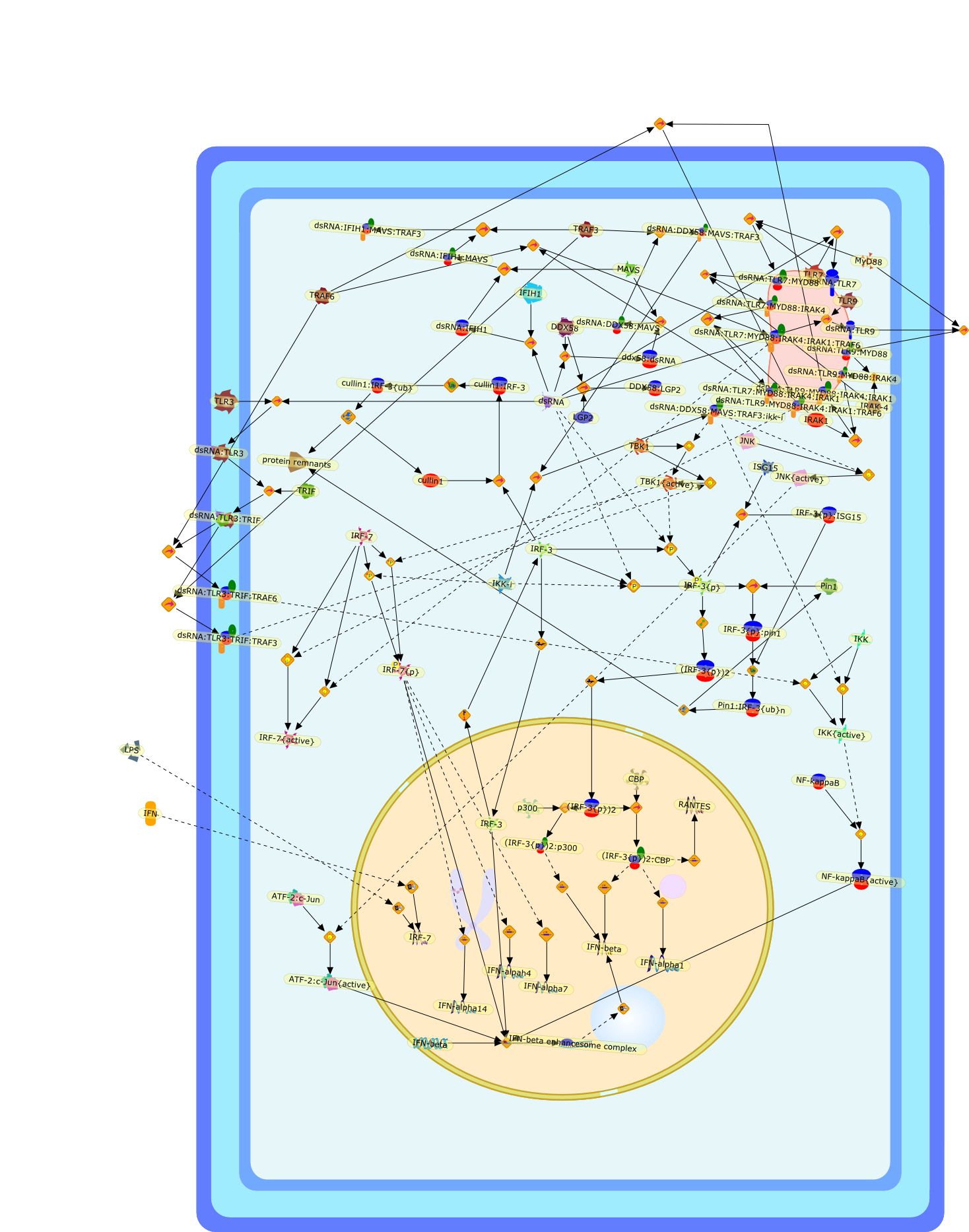
Triggering the innate antiviral response through IRF-3 activation.
Affiliation
Lady Davis Institute for Medical Research-Jewish General Hospital, Departmentsof Microbiology & Immunology, Medicine, and Oncology, McGill University,Montreal H3T 1E2, Canada. john.hiscott@mcgill.ca
Abstract
Rapid induction of type I interferon (IFN) expression is a central event in theestablishment of the innate immune response against viral infection and requiresthe activation of multiple transcriptional proteins following engagement andsignaling through Toll-like receptor-dependent and -independent pathways. Thetranscription factor interferon regulatory factor-3 (IRF-3) contributes to afirst line of defense against viral infection by inducing the production ofIFN-beta that in turn amplifies the IFN response and the development ofantiviral activity. In murine knock-out models, the absence of IRF-3 and theclosely related IRF-7 ablates IFN production and increases viral pathogenesis,thus supporting a pivotal role for IRF-3/IRF-7 in the development of the hostantiviral response.
PMID
17395583
|