| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Regulation of TLR4 signaling and the host interface with pathogens and danger:the role of RP105.
Affiliation
Division of Molecular Immunology, Cincinnati Children's Hospital Medical Centerand the University of Cincinnati College of Medicine, Cincinnati, Ohio 45229,USA.
Abstract
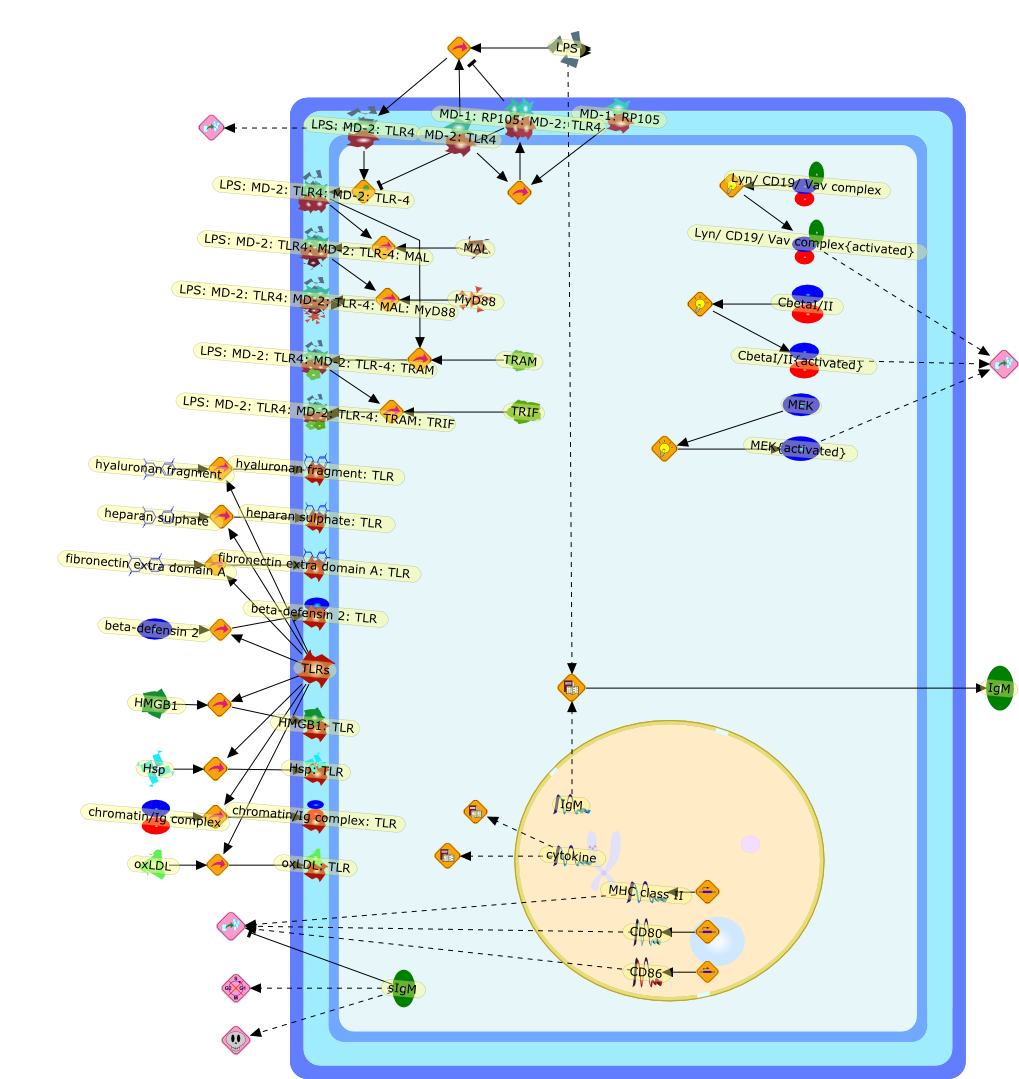
As all immune responses have potential for damaging the host, tight regulationof such responses--in amplitude, space, time and character--is essential formaintaining health and homeostasis. It was thus inevitable that the initial waveof papers on the role of Toll-like receptors (TLRs), NOD-like receptors (NLRs)and RIG-I-like receptors (RLRs) in activating innate and adaptive immuneresponses would be followed by a second wave of reports focusing on themechanisms responsible for restraining and modulating signaling by thesereceptors. This overview outlines current knowledge and controversies about theimmunobiology of the RP105/MD-1 complex, a modulator of the most robustlysignaling TLR, TLR4.
PMID
17470533
|