| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immuneresponse.
Affiliation
Department of Molecular Research and Development, Victorian Infectious DiseasesReference Laboratory, North Melbourne, Victoria, Australia.alexander.thompson@svhm.org.au
Abstract
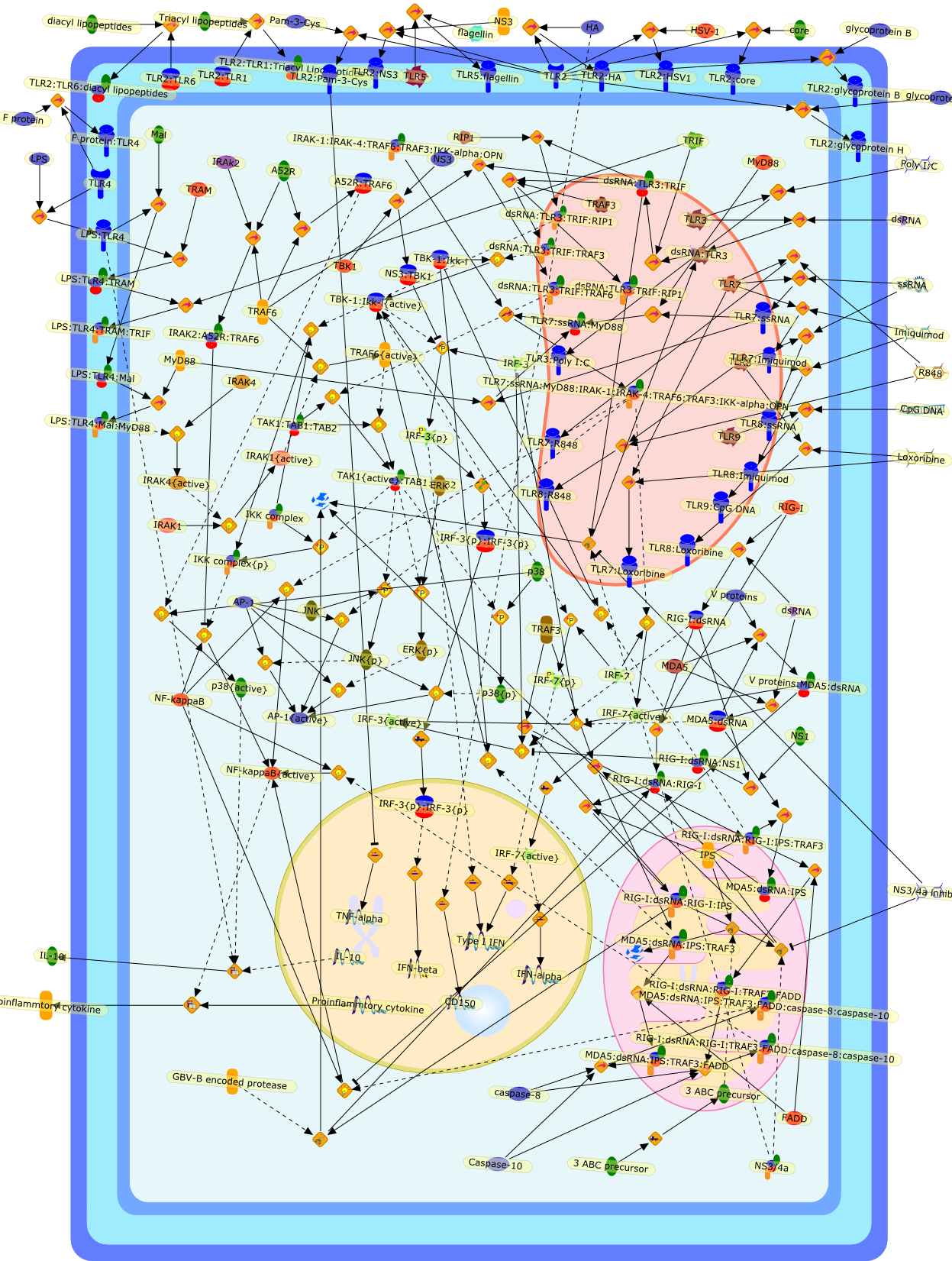
The antiviral innate immune response follows the detection of viral componentsby host pattern recognition receptors (PRRs). Two families of PRRs have emergedas key sensors of viral infection: Toll-like receptors (TLRs) and retinoic acidinducible gene-I like RNA helicases (RLHs). TLRs patrol the extracellular andendosomal compartments; signalling results in a type-1 interferon responseand/or the production of pro-inflammatory cytokines. In contrast, RLHs surveythe cytoplasm for the presence of viral double-stranded RNA. In the face of suchhost defence, viruses have developed strategies to evade TLR/RLH signalling.Such host-virus interactions provide the opportunity for manipulation of PRRsignalling as a novel therapeutic approach.
PMID
17667934
|