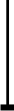| Original Literature | Model OverView |
|---|---|
|
Publication
Title
The role of the interferon regulatory factor (IRF) family in dendritic celldevelopment and function.
Affiliation
Department of Cell Biology and Neurosciences, Istituto Superiore di Sanita,Viale Regina Elena 299, 00161 Roma, Italy. lucia.gabriele@iss.it
Abstract
Dendritic cells (DCs) are powerful sensors of foreign pathogens as well ascancer cells and provide the first line of defence against infection. They alsoserve as a major link between innate and adaptive immunity. Immature DCs respondto incoming danger signals and undergo maturation to produce high levels ofproinflammatory cytokines including type I interferons (IFNs) to establishinnate immunity. They then present antigens to T lymphocytes to stimulatelasting specific immune responses. Recent studies point to the importance of DCsin the induction of peripheral tolerance. Transcription factors of the IRFfamily have emerged as crucial controllers of many aspects of DC activity,playing an essential role in the establishment of early innate immunity.Furthermore, eight of the nine members of the IRF family have been shown tocontrol either the differentiation and/or the functional activities of DCs. Inthis review, we focus on three aspects of DC properties that are under thecontrol of IRFs: (1) the development and differentiation, (2) maturation inresponse to toll-like receptor (TLR) signalling and the production ofanti-microbial cytokines, and (3) activation and expansion of lymphocytes togenerate protective or tolerogenic immune responses.
PMID
17702640
|