| Original Literature | Model OverView |
|---|---|
|
Publication
Title
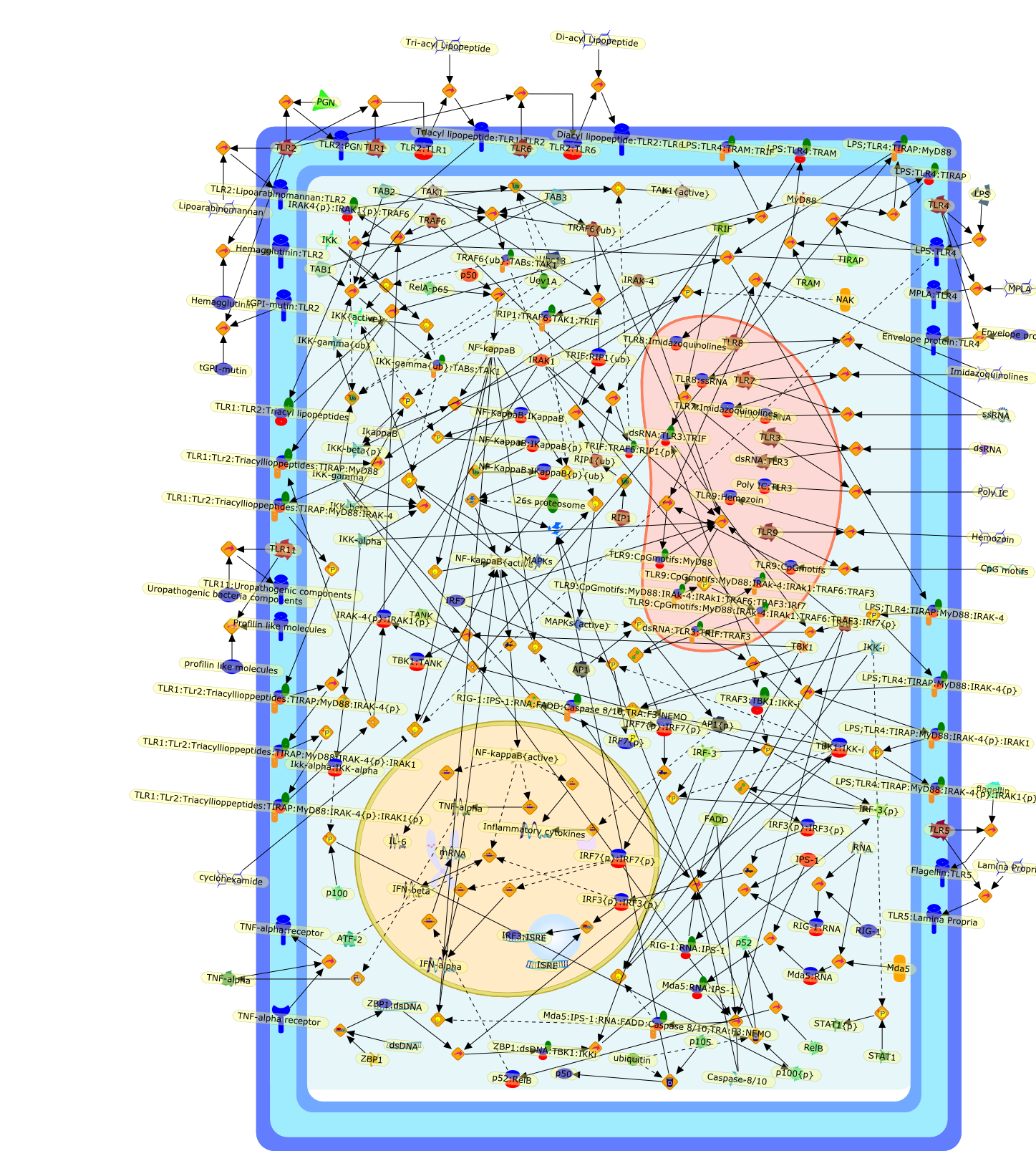
Signaling to NF-kappaB by Toll-like receptors.
Affiliation
Department of Host Defense and Exploratory Research for Advanced Technology,Japan Science and Technology Agency, Research Institute for Microbial Diseases,Osaka University, 3-1 Yamada-oka, Suita, Osaka 565-0871, Japan.
Abstract
Innate immunity is the first line of defense against invading pathogens. Afamily of Toll-like receptors (TLRs) acts as primary sensors that detect a widevariety of microbial components and elicit innate immune responses. All TLRsignaling pathways culminate in activation of the transcription factor nuclearfactor-kappaB (NF-kappaB), which controls the expression of an array ofinflammatory cytokine genes. NF-kappaB activation requires the phosphorylationand degradation of inhibitory kappaB (IkappaB) proteins, which is triggered bytwo kinases, IkappaB kinase alpha (IKKalpha) and IKKbeta. In addition, severalTLRs activate alternative pathways involving the IKK-related kinases TBK1 [TRAFfamily member-associated NF-kappaB activator (TANK) binding kinase-1] and IKKi,which elicit antiviral innate immune responses. Here, we review recent progressin our understanding of the role of NF-kappaB in TLR signaling pathways anddiscuss potential implications for molecular medicine.
PMID
18029230
|