| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated?
Affiliation
Interdisciplinary Cluster for Applied Genoproteomics, University of Liege,Sart-Tilman, 4000 Liege, Belgium.
Abstract
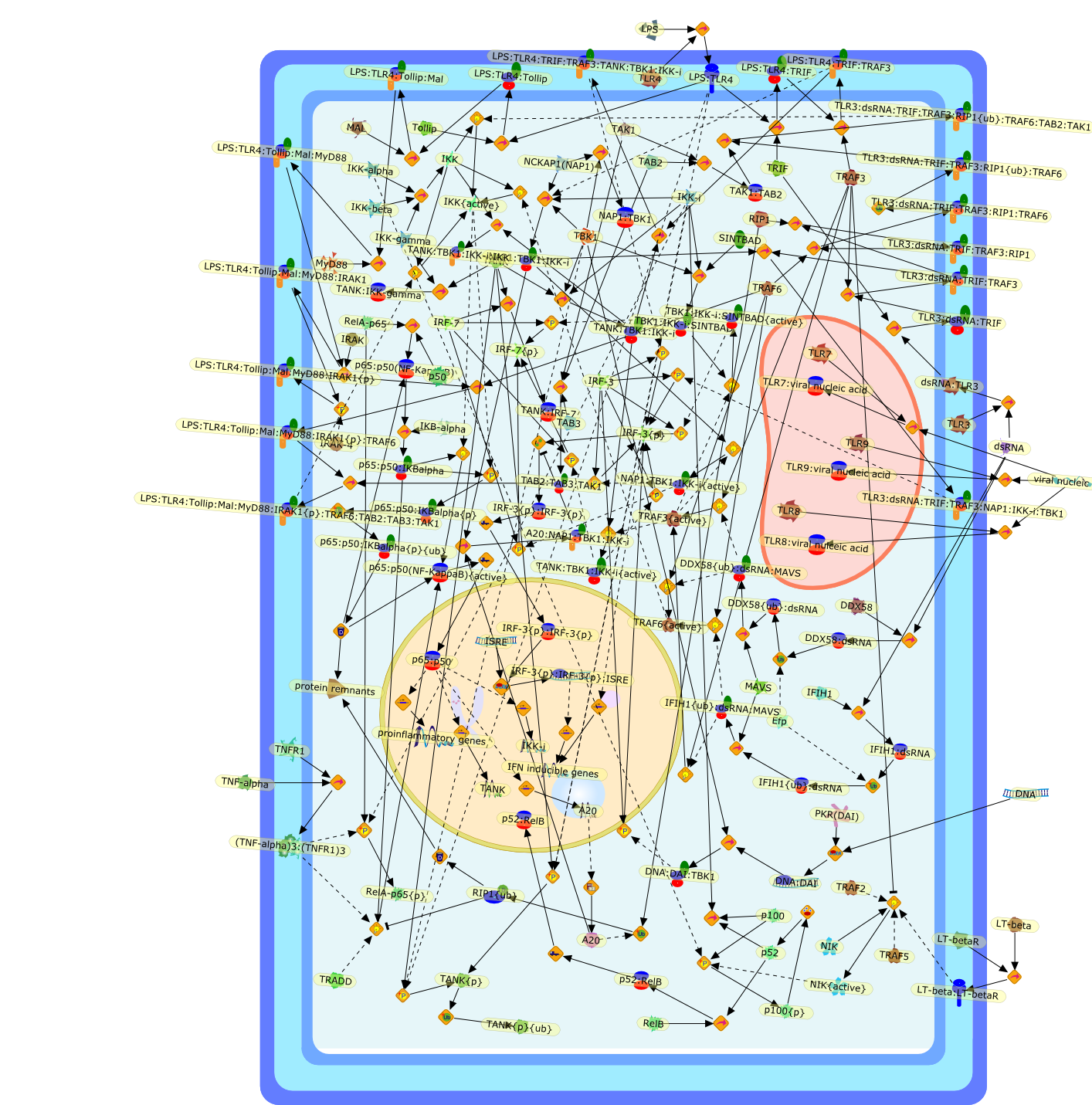
The IkappaB kinases (IKKs) IKK-alpha and IKK-beta, and the IKK-related kinasesTBK1 and IKK-epsilon, have essential roles in innate immunity throughsignal-induced activation of NF-kappaB, IRF3 and IRF7, respectively. Althoughthe signaling events within these pathways have been extensively studied, themechanisms of IKK and IKK-related complex assembly and activation remain poorlydefined. Recent data provide insight into the requirement for scaffold proteinsin complex assembly; NF-kappaB essential modulator coordinates some IKKcomplexes, whereas TANK, NF-kappaB-activating kinase-associated protein 1 (NAP1)or similar to NAP1 TBK1 adaptor (SINTBAD) assemble TBK1 and IKK-epsiloncomplexes. The different scaffold proteins undergo similar post-translationalmodifications, including phosphorylation and non-degradative polyubiquitylation.Moreover, increasing evidence indicates that distinct scaffold proteins assembleIKK, and potentially TBK1 and IKK-epsilon subcomplexes, in a stimulus-specificmanner, which might be a mechanism to achieve specificity.
PMID
18353649
|