| Original Literature | Model OverView |
|---|---|
|
Publication
Title
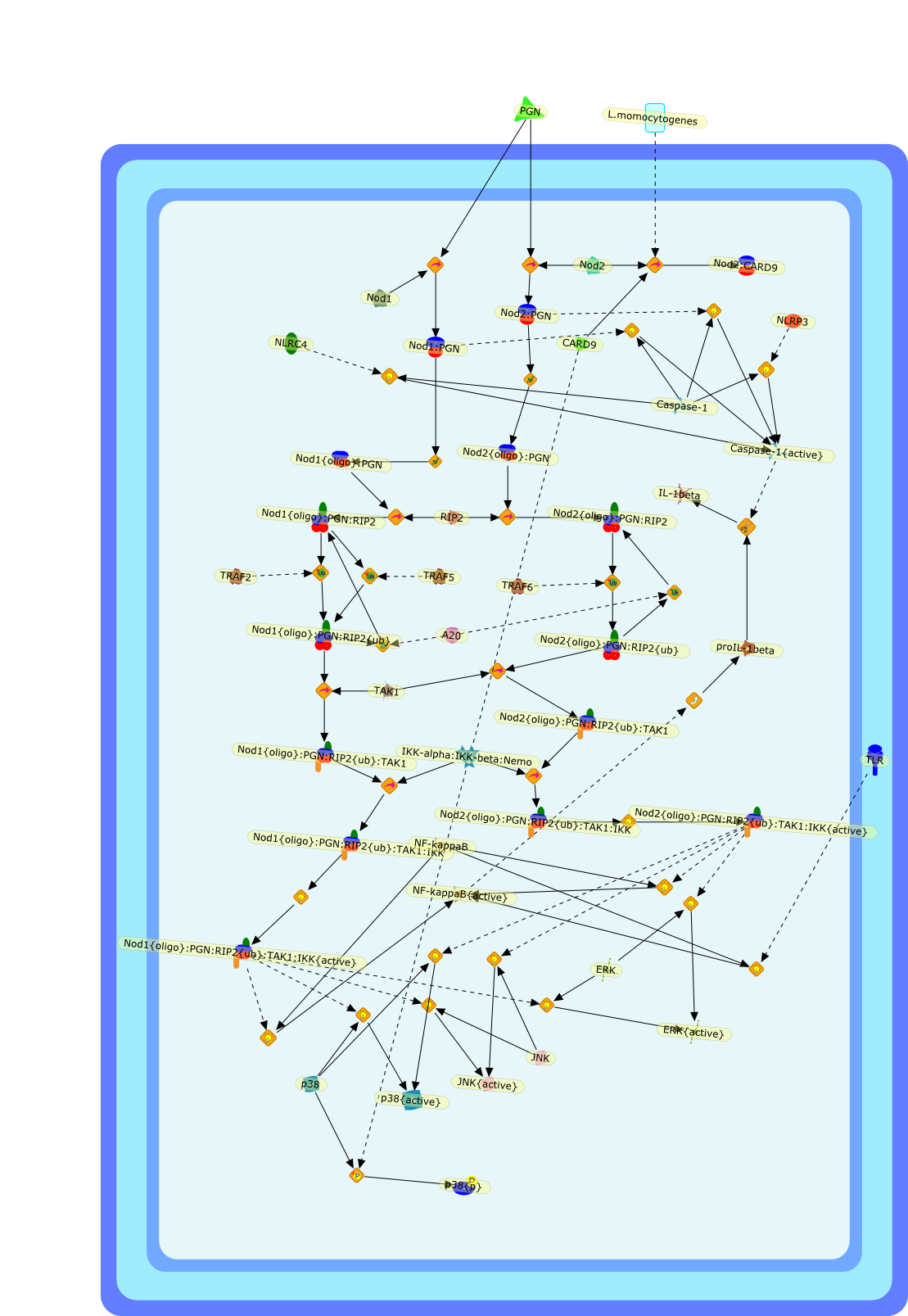
NOD-like receptors (NLRs): bona fide intracellular microbial sensors.
Affiliation
Department of Pathology and Comprehensive Cancer Center, University of MichiganMedical School, Ann Arbor, MI 48109, USA.
Abstract
The nucleotide-binding oligomerization domain (NOD)-like receptor (NLR)(nucleotide-binding domain leucine-rich repeat containing) family of proteinshas been demonstrated to function as regulators of innate immune responseagainst microbial pathogens. Stimulation of NOD1 and NOD2, two prototypic NLRs,results in the activation of MAPK and NF-kappaB. On the other hand, a differentset of NLRs induces caspase-1 activation through the assembly of aninflammasome. This review discusses recent findings regarding the signalingpathways utilized by NLR proteins in the control of caspase-1 and NF-kappaBactivation, as well as the nonredundant role of NLRs in pathogen clearance. Thereview also covers advances regarding the cellular localization of theseproteins and the implications this may have on pathogen sensing and signaltransduction.
PMID
18585455
|