| Original Literature | Model OverView |
|---|---|
|
Publication
Title
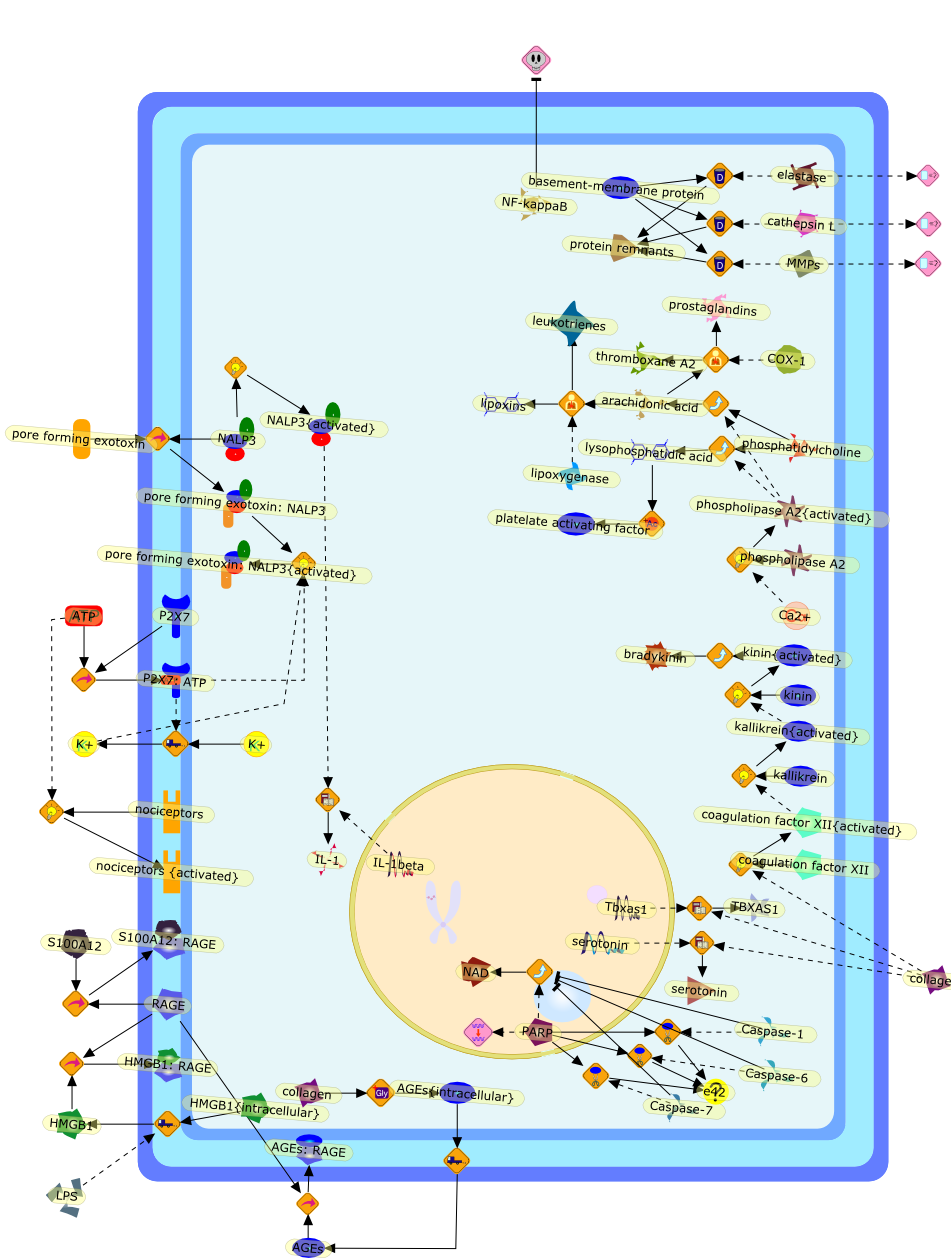
Origin and physiological roles of inflammation.
Affiliation
Howard Hughes Medical Institute and Department of Immunobiology, Yale UniversitySchool of Medicine, TAC S-669, 300 Cedar Street, New Haven, Connecticut 06510,USA. ruslan.medzhitov@yale.edu
Abstract
Inflammation underlies a wide variety of physiological and pathologicalprocesses. Although the pathological aspects of many types of inflammation arewell appreciated, their physiological functions are mostly unknown. The classicinstigators of inflammation - infection and tissue injury - are at one end of alarge range of adverse conditions that induce inflammation, and they trigger therecruitment of leukocytes and plasma proteins to the affected tissue site.Tissue stress or malfunction similarly induces an adaptive response, which isreferred to here as para-inflammation. This response relies mainly ontissue-resident macrophages and is intermediate between the basal homeostaticstate and a classic inflammatory response. Para-inflammation is probablyresponsible for the chronic inflammatory conditions that are associated withmodern human diseases.
PMID
18650913
|