| Original Literature | Model OverView |
|---|---|
|
Publication
Title
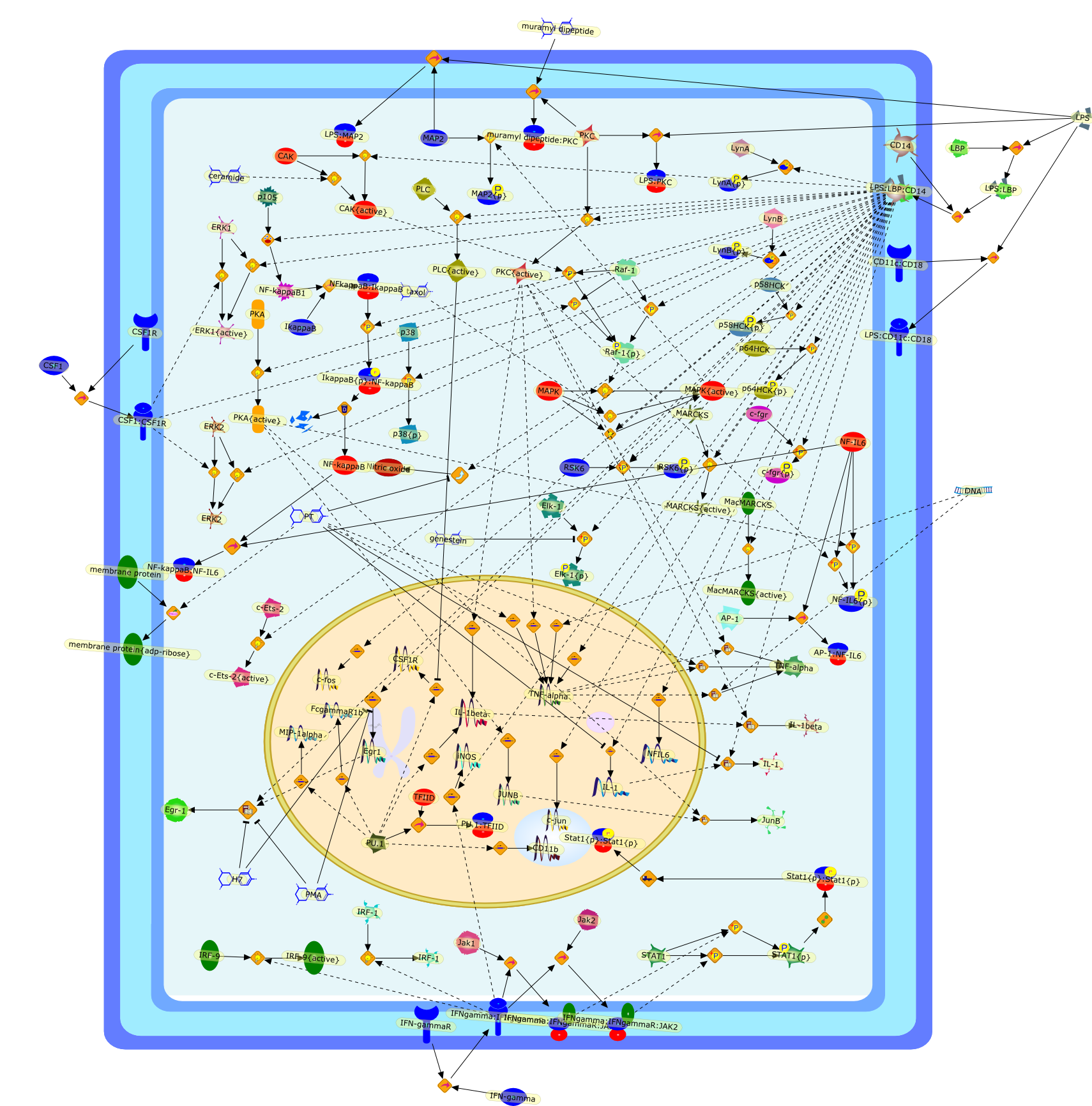
Endotoxin signal transduction in macrophages.
Affiliation
Centre for Molecular and Cellular Biology, University of Queensland, Brisbane,Australia.
Abstract
Through its action on macrophages, bacterial lipopolysaccharide (LPS) orendotoxin can trigger responses that are protective or injurious to the host.This review examines the effects of LPS on macrophages by following events fromthe cell surface to the nucleus. The involvement of protein tyrosine kinases,mitogen-activated protein kinases, protein kinase C, G proteins, protein kinaseA, ceramide-activated protein kinase, and microtubules in this process arereviewed. At the nuclear level, rel, C/EBP, Ets, Egr, fos, and jun familymembers have been implicated in activation of LPS-inducible gene expression.
PMID
8699127
|