| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Lipoprotein trafficking in vascular cells. Molecular Trojan horses and cellularsaboteurs.
Affiliation
Cornell University Medical College, New York, New York 10021 and UCLA School ofMedicine, Los Angeles, California 90095, USA.
Abstract
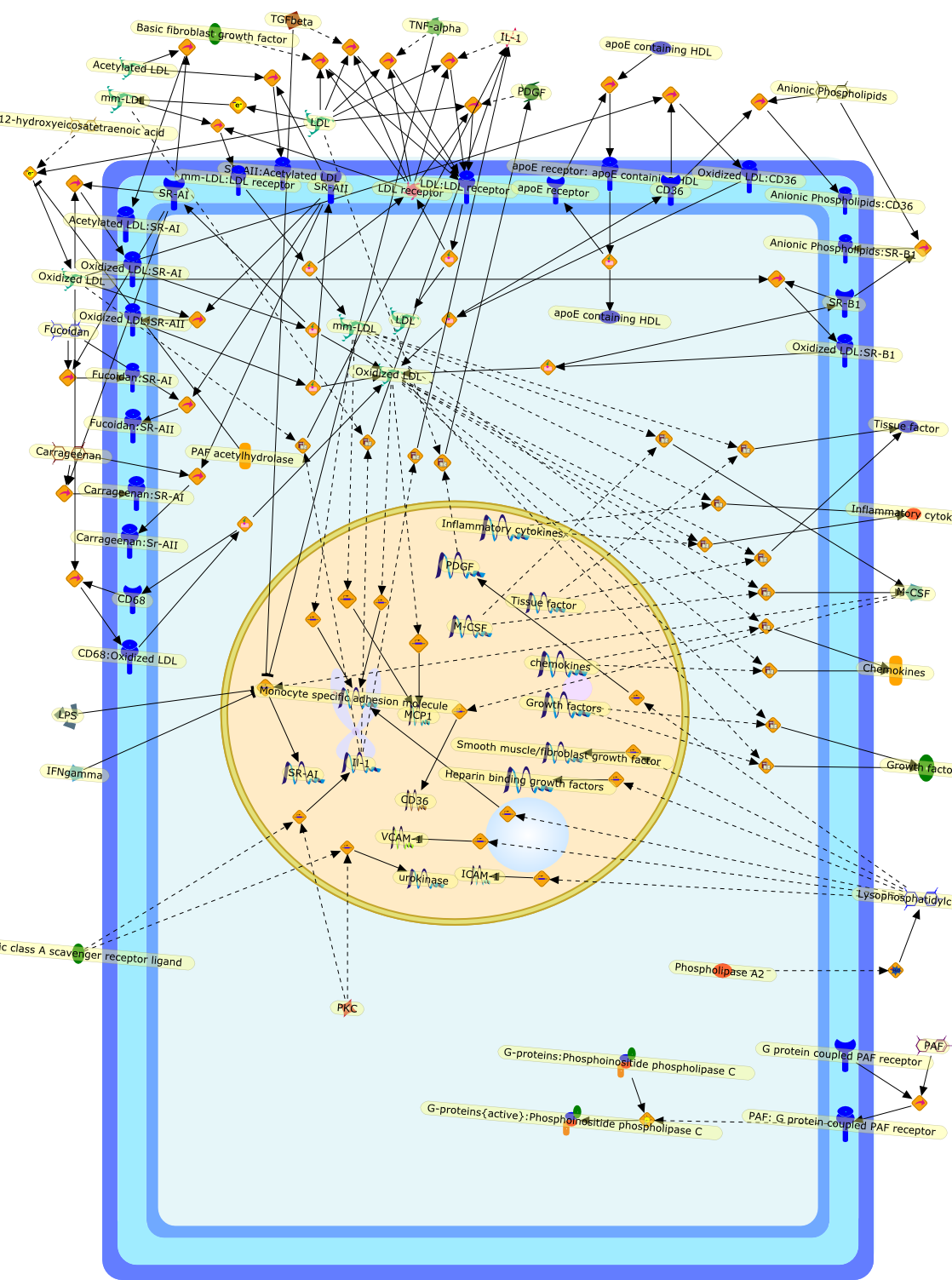
During the pathogenesis of atherosclerosis, inflammatory cells such as themonocyte-derived macrophage accumulate in the vessel wall where they releasecytokines. Initially, cytokines may assist in CE removal of lipoprotein-derivedcholesterol/CE hydrolysis to clear intracellular lipid. When plasma levels ofLDL become elevated, the vessel wall becomes lipid-engorged over time because itis unable to traffick the large amounts of endocytosed LDL-CE from the cell. Inaddition, lipoprotein entrapment by the extracellular matrix can lead to theprogressive oxidation of LDL because of the action of lipoxygenases, reactiveoxygen species, peroxynitrite, and/or myeloperoxidase. A range of oxidized LDLspecies is thus generated, ultimately resulting in their delivery to vascularcells through several families of scavenger receptors (Fig 1). These molecularTrojan horses and cellular saboteurs once formed or deposited in the cell cancontribute to, and participate in, formation of macrophage- and smoothmuscle-derived foam cells. A lipid-enriched fatty streak along the vessel wallcan ensue. In addition to foam cell development, products of LDL peroxidationmay activate endothelial cells, increase smooth muscle mitogenesis, or induceapoptosis because of the effects of oxysterols and products of lipidperoxidation (Fig 1). Because antioxidant defenses may be limited in themicroenvironment of the cell or within LDL, the oxidation process continues toprogress. Enzymes associated with HDL such as PAF acetylhydrolase andparaoxonase can participate in the elimination of biologically active lipids,but diminished cellular antioxidant activity coupled with low levels of HDL mayallow acceleration of the clinical course of vascular disease. There is stillmuch to be learned about how modified LDL initiate cellular signals that lead toinflammation, mitosis, or cholesterol accumulation. The present challengesinclude elucidation of the key signaling events that regulatelipoprotein-derived cholesterol trafficking in the vessel wall, which can impacton the pathogenesis of vascular disease.
PMID
9287290
|