| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Differential responses of human monocytes and macrophages to IL-4 and IL-13.
Affiliation
Department of Microbiology & Infectious Diseases, School of Medicine, FlindersUniversity of South Australia, Adelaide, Australia.
Abstract
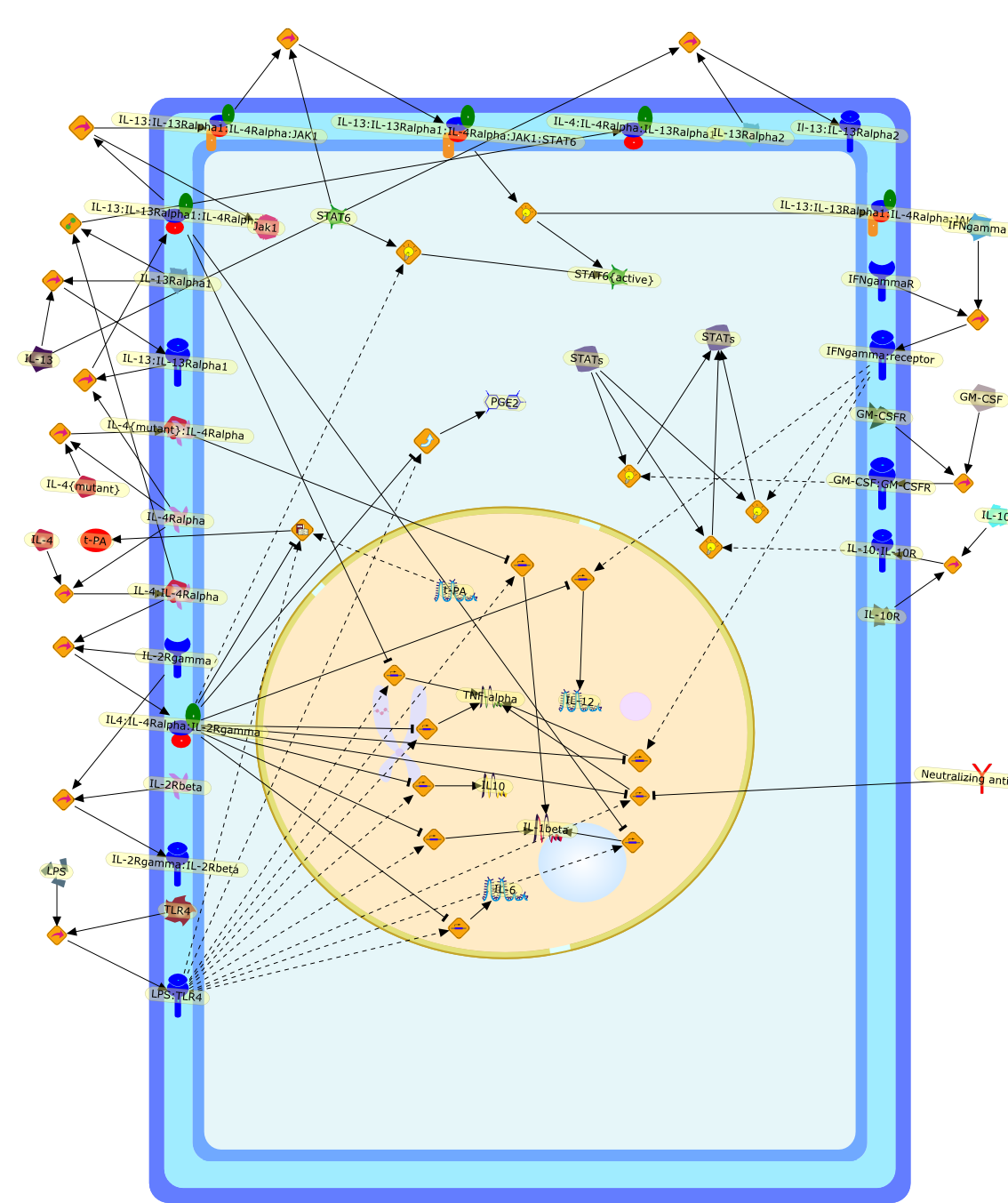
The primary interleukin-4 (IL-4) receptor complex on monocytes (type I IL-4receptor) includes the 140-kDa alpha chain (IL-4R alpha) and the IL-2 receptorgamma chain, gamma(c), which heterodimerize for intracellular signaling,resulting in suppression of lipopolysaccharide (LPS)-inducible inflammatorymediator production. The activity of IL-13 on human monocytes is very similar tothat of IL-4 because the predominant signaling chain (IL-4R alpha) is common toboth receptors. In fact, IL-4R alpha with IL-13R alpha1 is designated both as anIL-13 receptor and the type II IL-4 receptor. When the anti-inflammatoryactivities of IL-4 and IL-13 were investigated on synovial fluid macrophages andcompared with the responses by monocytes isolated from the patients at the sametime as joint drainage, the response profiles differed with some responsessimilar in the two cell populations, others reduced on the inflammatory cells.Similar differences were recorded in the response profiles to IL-4 and IL-13 bymonocytes and monocytes cultured for 7 days in macrophage colony-stimulatingfactor (M-CSF) or granulocyte-macrophage CSF (GM-CSF) (monocyte-derivedmacrophages, MDMac). MDMac have reduced gamma(c) mRNA levels and reducedexpression of the functional 64-kDa gamma(c). There was a similar loss of IL-13Ralpha1 mRNA on monocyte differentiation. In turn, there was a significantreduction in the ability of IL-4 and IL-13 to activate STAT6. These findingssuggest that different functional responses to IL-4 and IL-13 by human monocytesand macrophages may result from reduced expression of gamma(c) and IL-13Ralpha1.
PMID
10534111
|