| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Pathophysiological roles of interleukin-18 in inflammatory liver diseases.
Affiliation
Department of Immunology and Medical Zoology, Hyogo College of Medicine,Nishinomiya, Japan.
Abstract
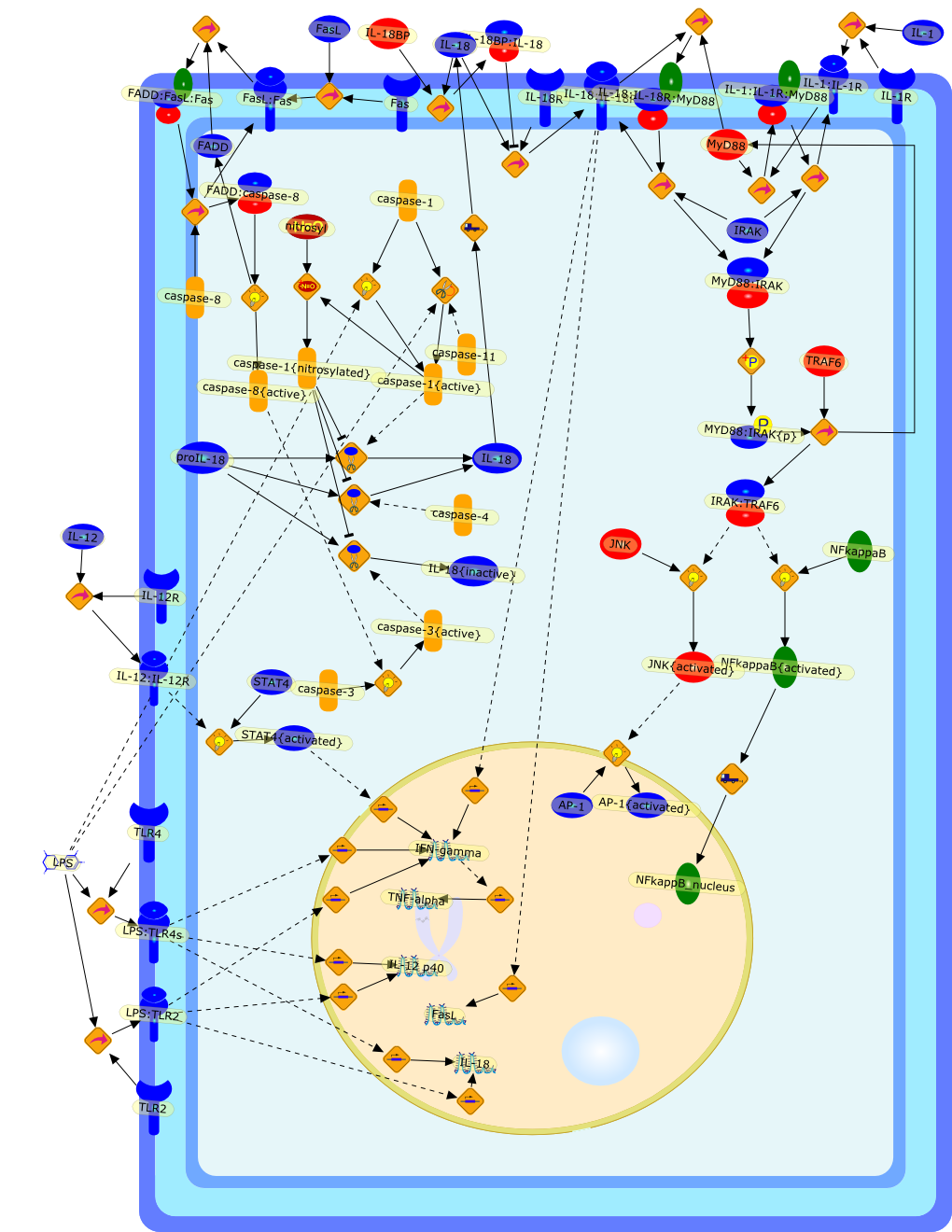
Innate immune response to microbes sometimes determines the nature of thefollowing specific immune response. Kupffer cells, a potent constituent ofinnate immunity, play a key role in developing the type 1 immune response byinterleukin (IL)-12 production. Furthermore, Kupffer cells have the potential toinduce liver injury by production of IL-18. Propionibacterium acnes-primedlipopolysaccharide (LPS)-challenged liver injury is the prototype ofIL-18-induced tissue injury, in which IL-18 acts on natural killer cells toincrease Fas ligand (FasL) that causes liver injury by induction ofFas-dependent hepatocyte apoptosis. LPS induces IL-18 secretion from Kupffercells in a caspase-1-dependent manner. Indeed, caspase-1-deficient mice areresistant to P. acnes and LPS-induced liver injury. However, administration ofsoluble FasL induces acute liver injury in P. acnes-primed caspase-1-deficientmice but does not do so in IL-18-deficient mice, indicating that IL-18 releasein a caspase-1-independent fashion is essential for this liver injury.Therefore, a positive feedback loop between FasL and IL-18 plays an importantrole in the pathogenesis of endotoxin-induced liver injury.
PMID
10807517
|