| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Pathogen-induced apoptosis of macrophages: a common end for different pathogenicstrategies.
Affiliation
Skirball Institute for Biomolecular Medicine, Department of Microbiology andKaplan Cancer Center, New York University School of Medicine, NY 10028, USA.
Abstract
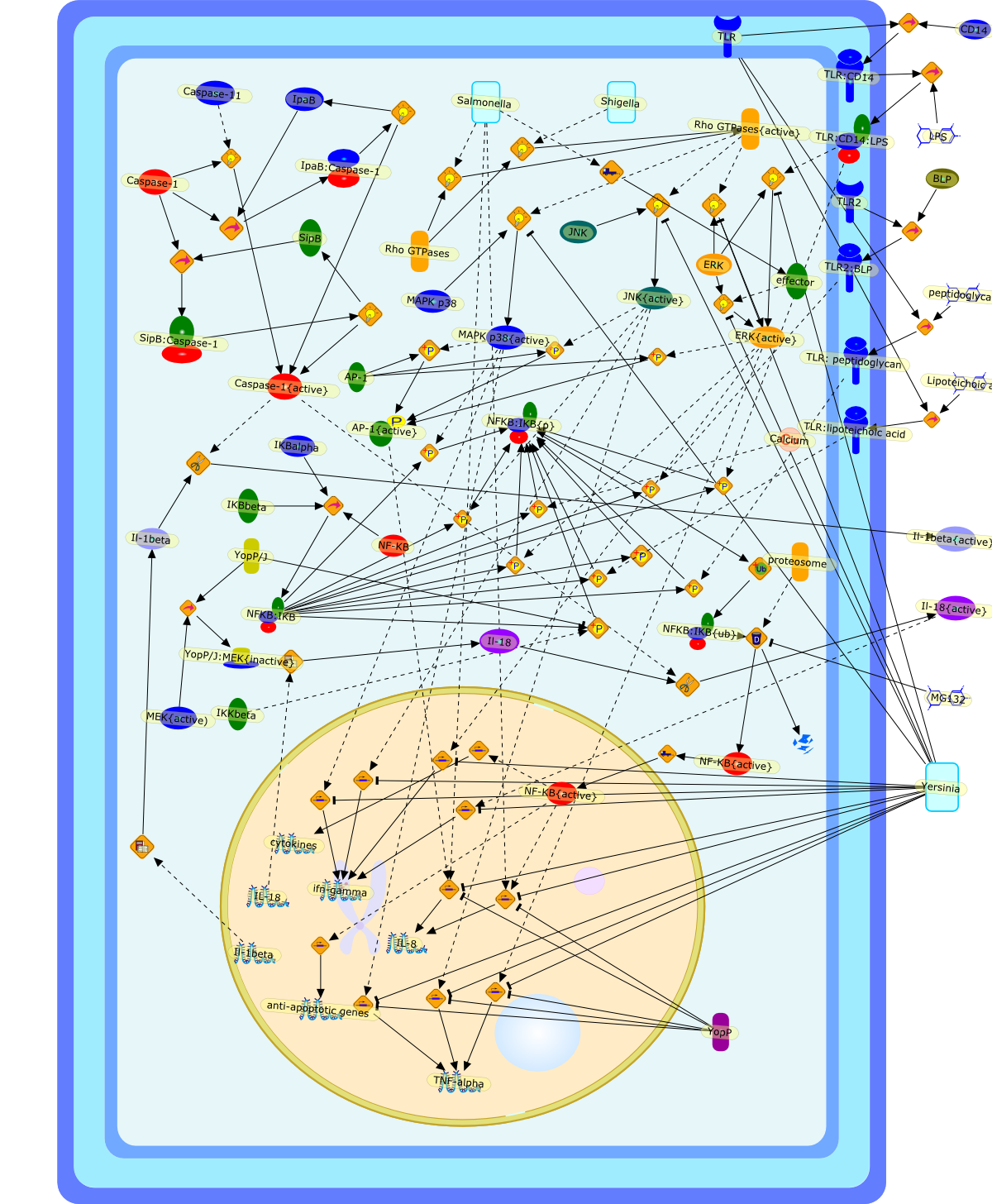
Microbe-macrophage interactions play a central role in the pathogenesis of manyinfections. Several bacterial pathogens induce apoptosis specifically inmacrophages, but the mechanisms by which it occurs differ, and the resultingpathology can take different courses. Macrophage death caused by Shigellaflexneri and Salmonella spp. has been shown to result in the release ofpro-inflammatory cytokines. Conversely, Yersinia spp. induce apoptosis bysuppressing the signalling pathways that lead to the production of tumournecrosis factor (TNF)-alpha, a cytokine essential for the control of thisinfection. It is likely that there are a variety of reasons why macrophages areparticularly susceptible to pathogen-induced apoptosis. One reason may be theexpression of surface receptors that recognize highly conserved bacterialcomponents, such as lipopolysaccharide (LPS) and bacterial lipoproteins (BLPs).These receptors have recently been shown to activate pro-apoptotic signallingpathways. The roles of macrophage apoptosis in different disease processes arediscussed.
PMID
11207583
|