| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Molecular aspects of anti-inflammatory action of G-CSF.
Affiliation
Biochemical Pharmacology, University of Konstanz, Germany.
Abstract
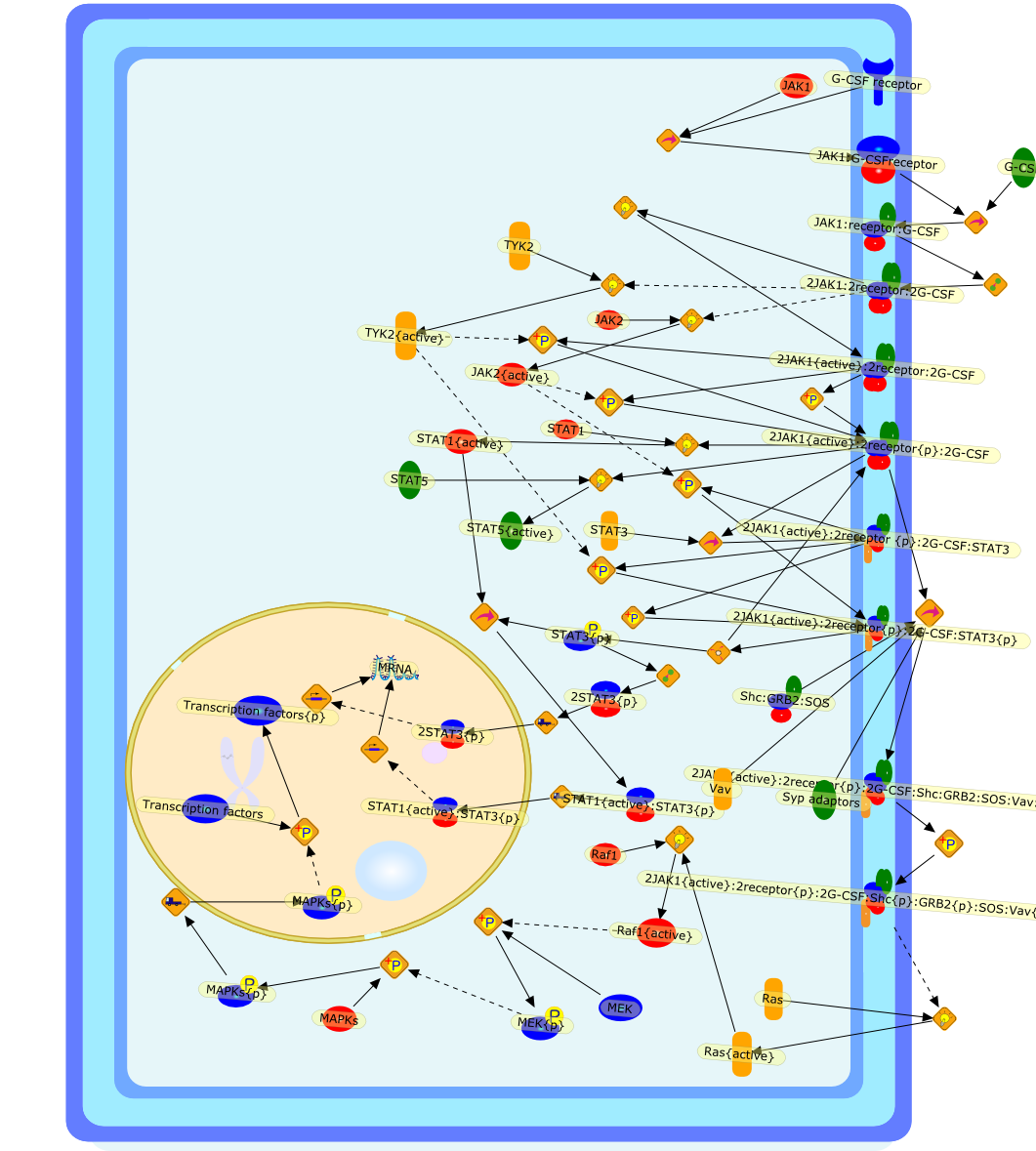
Granulocyte colony-stimulating factor (G-CSF) is a cytokine which stimulates theproduction of neutrophils in the bone marrow and modulates cellular functions ofmature neutrophils. Besides neutrophils and their precursors, monocytes aredirect target cells of G-CSF action. G-CSF influences monocyte functions in ananti-inflammatory way: The stimulation of monocytes with G-CSF results in anattenuation of LPS-induced release of IL-1beta, TNF-alpha, IL-12 and IL-18.G-CSF exerts its biological functions on neutrophils and monocytes viamembrane-bound receptors. Seven different human G-CSF receptor isoforms havebeen described which are generated by alternative splicing. The physiologicroles of these isoforms and the expression pattern on various cell types arestill unknown. The signal transduction pathway of G-CSF receptors involves theactivation of JAK tyrosine kinases and STAT transcription factors and theras/mitogen-activated protein kinase pathway.
PMID
12005202
|