| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Structural and functional analyses of bacterial lipopolysaccharides.
Affiliation
Equipe Endotoxines, UMR 8619 du Centre National de la Recherche Scientifique,Biochimie, Universite de Paris-Sud, Orsay, France.martine.caroff@bbmpc.u-psud.fr
Abstract
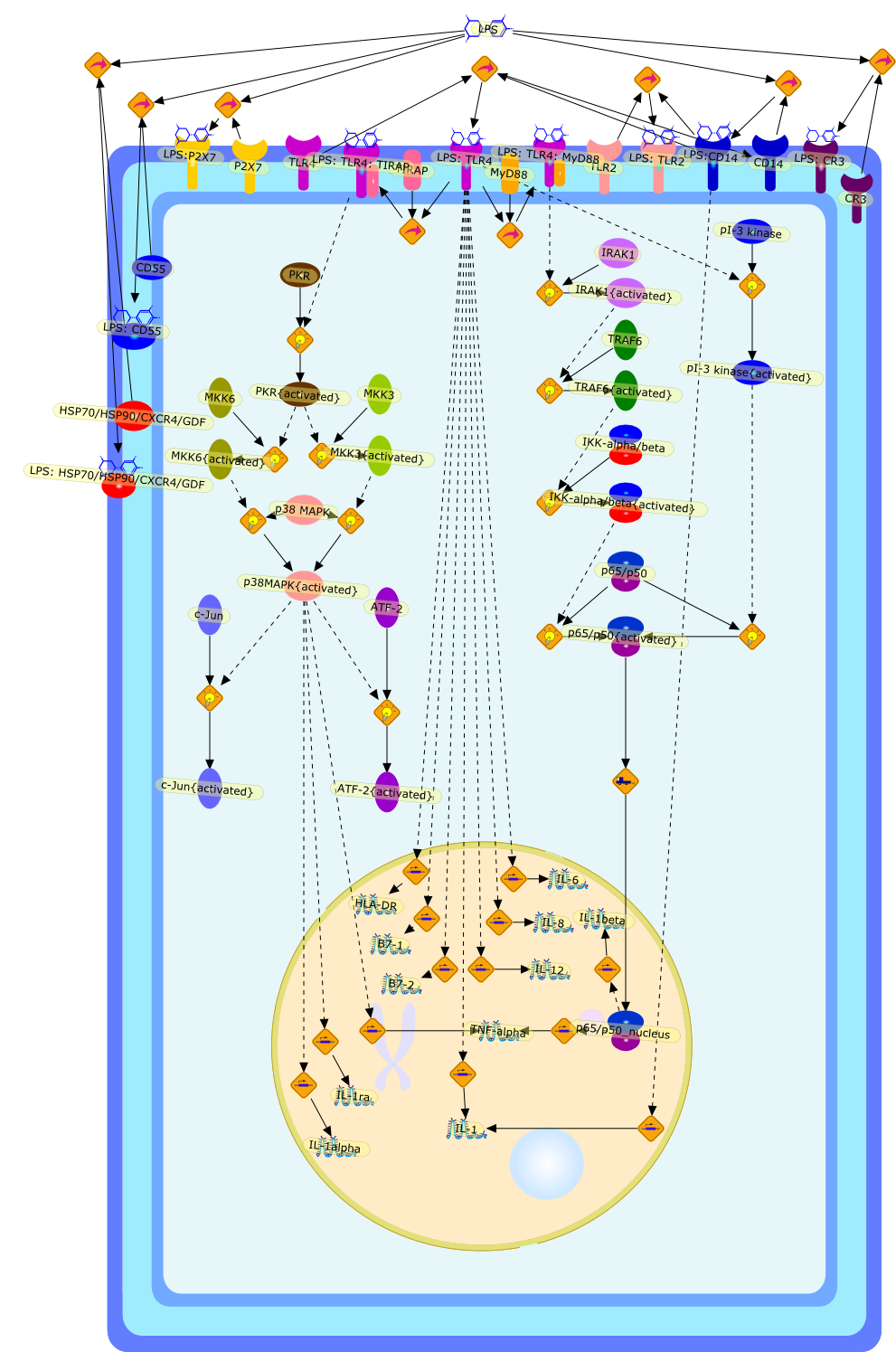
Bacterial lipopolysaccharides (LPSs) are powerful immunomodulators in infectedhosts, and may cause endotoxic shock. Most of them share a common architecturebut vary considerably in structural motifs from one genus, species, and strainto another. Cells of the innate immune response recognize evolutionarilyconserved LPS molecular patterns of endotoxins and structural details therebygreatly influencing their response.
PMID
12106784
|