| Original Literature | Model OverView |
|---|---|
|
Publication
Title
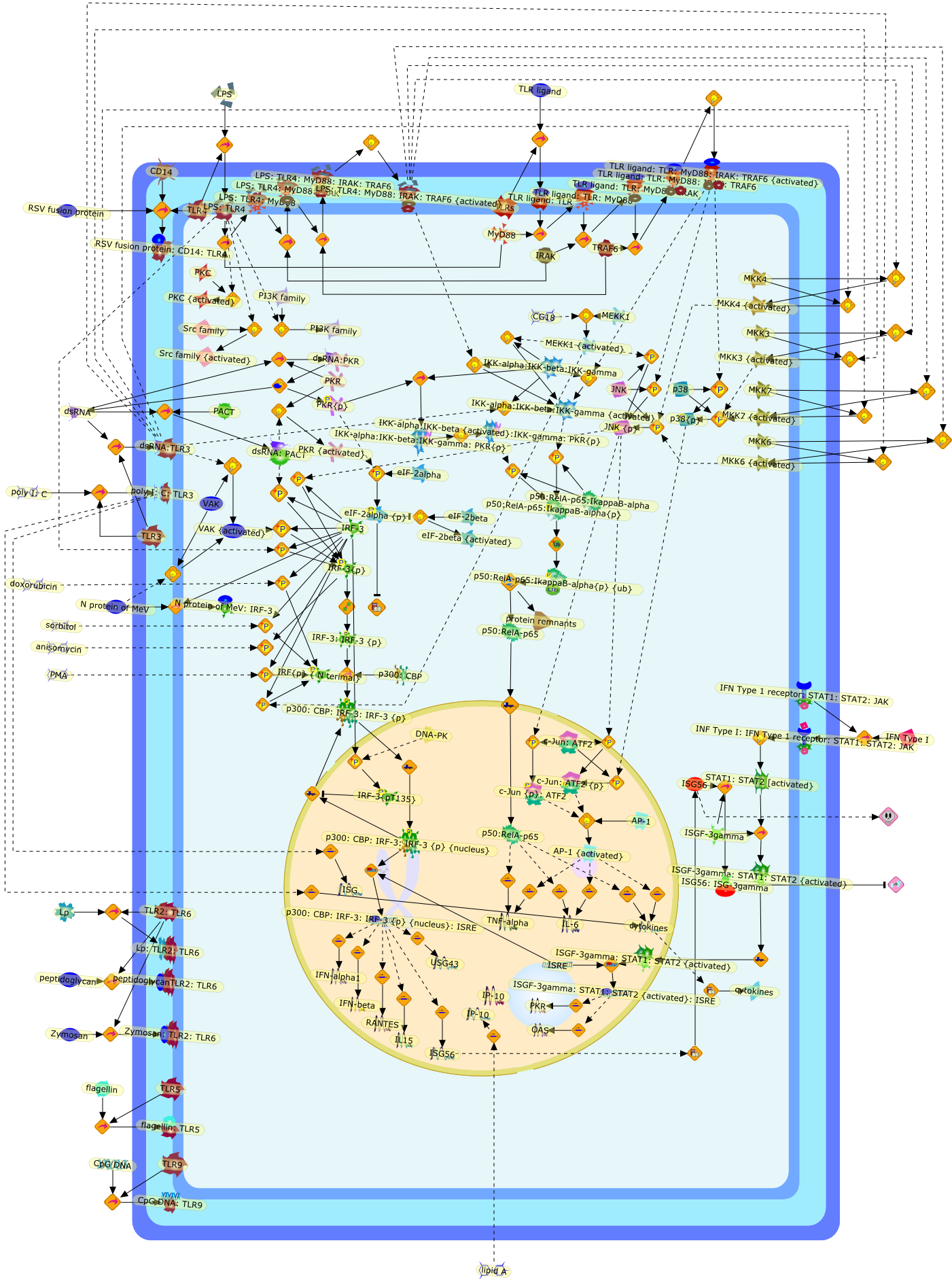
Multiple signaling pathways leading to the activation of interferon regulatoryfactor 3.
Affiliation
Terry Fox Molecular Oncology Group, Lady Davis Institute-Jewish GeneralHospital, McGill University, 3755 Cote Ste., Catherine Montreal, Que., Canada.
Abstract
Virus infection of susceptible cells activates multiple signaling pathways thatorchestrate the activation of genes, such as cytokines, involved in theantiviral and innate immune response. Among the kinases induced are themitogen-activated protein (MAP) kinases, Jun-amino terminal kinases (JNK) andp38, the IkappaB kinase (IKK) and DNA-PK. In addition, virus infection alsoactivates an uncharacterized VAK responsible for the C-terminal phosphorylationand subsequent activation of interferon regulatory factor 3 (IRF-3).Virus-mediated activation of IRF-3 through VAK is dependent on viral entry andtranscription, since replication deficient virus failed to induce IRF-3activity. The pathways leading to VAK activation are not well characterized, butIRF-3 appears to represent a novel cellular detection pathway that recognizesviral nucleocapsid (N) structure. Recently, the range of inducers responsiblefor IRF-3 activation has increased. In addition to virus infection, recognitionof bacterial infection mediated through lipopolysaccharide by Toll-like receptor4 has also been reported. Furthermore, MAP kinase kinase kinase (MAPKKK)-related pathways and DNA-PK induce N-terminal phosphorylation of IRF-3.This review summarizes recent observations in the identification of novelsignaling pathways leading to IRF-3 activation.
PMID
12213596
|