| Original Literature | Model OverView |
|---|---|
|
Publication
Title
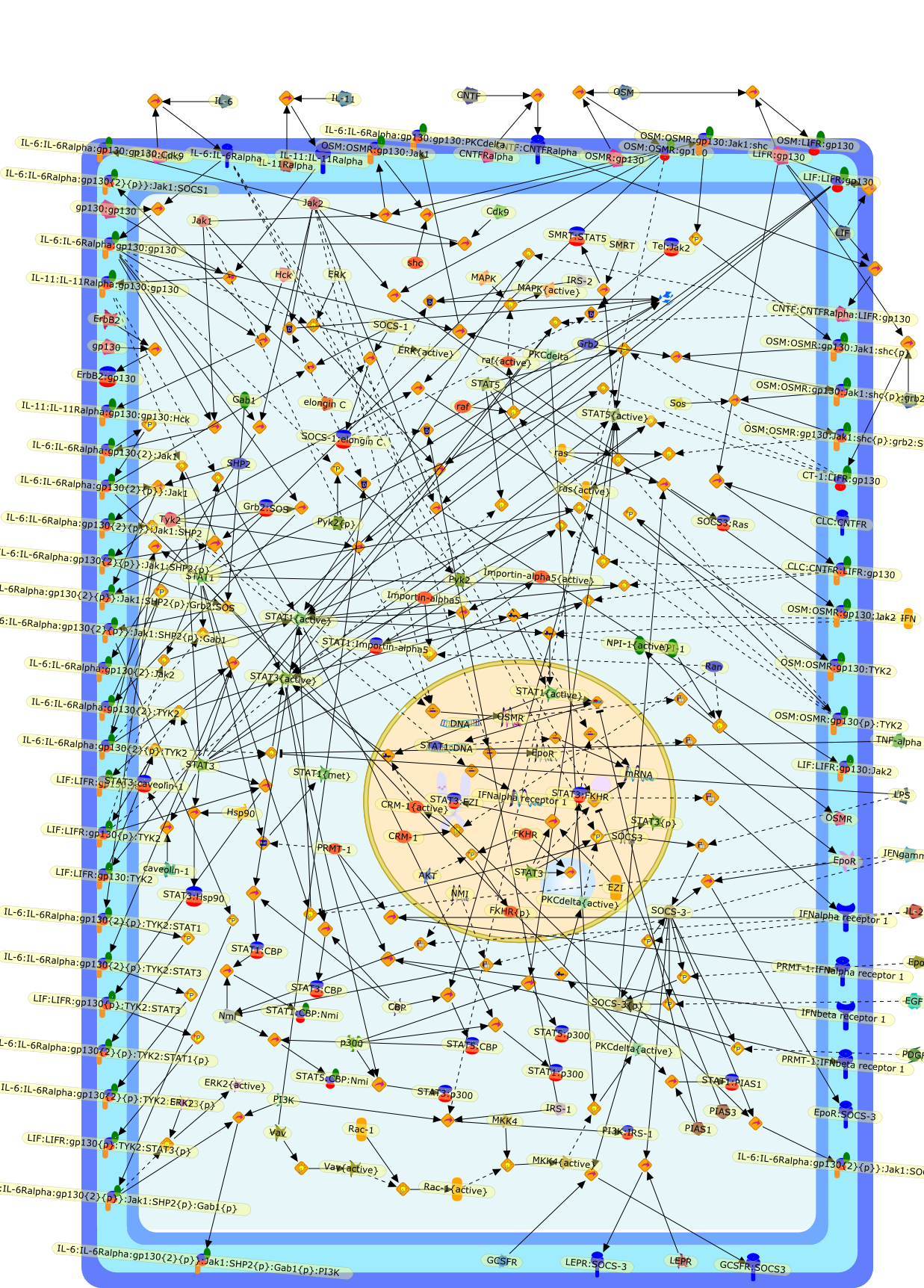
Principles of interleukin (IL)-6-type cytokine signalling and its regulation.
Affiliation
Institut fur Biochemie, RWTH Aachen, Universitatsklinikum, Pauwelsstrasse 30,D-52074 Aachen, Germany. heinrich@rwth-aachen.de
Abstract
The IL (interleukin)-6-type cytokines IL-6, IL-11, LIF (leukaemia inhibitoryfactor), OSM (oncostatin M), ciliary neurotrophic factor, cardiotrophin-1 andcardiotrophin-like cytokine are an important family of mediators involved in theregulation of the acute-phase response to injury and infection. Besides theirfunctions in inflammation and the immune response, these cytokines play also acrucial role in haematopoiesis, liver and neuronal regeneration, embryonaldevelopment and fertility. Dysregulation of IL-6-type cytokine signallingcontributes to the onset and maintenance of several diseases, such as rheumatoidarthritis, inflammatory bowel disease, osteoporosis, multiple sclerosis andvarious types of cancer (e.g. multiple myeloma and prostate cancer). IL-6-typecytokines exert their action via the signal transducers gp (glycoprotein) 130,LIF receptor and OSM receptor leading to the activation of the JAK/STAT (Januskinase/signal transducer and activator of transcription) and MAPK(mitogen-activated protein kinase) cascades. This review focuses on recentprogress in the understanding of the molecular mechanisms of IL-6-type cytokinesignal transduction. Emphasis is put on the termination and modulation of theJAK/STAT signalling pathway mediated by tyrosine phosphatases, the SOCS(suppressor of cytokine signalling) feedback inhibitors and PIAS (proteininhibitor of activated STAT) proteins. Also the cross-talk between the JAK/STATpathway with other signalling cascades is discussed.
PMID
12773095
|