| Original Literature | Model OverView |
|---|---|
|
Publication
Title
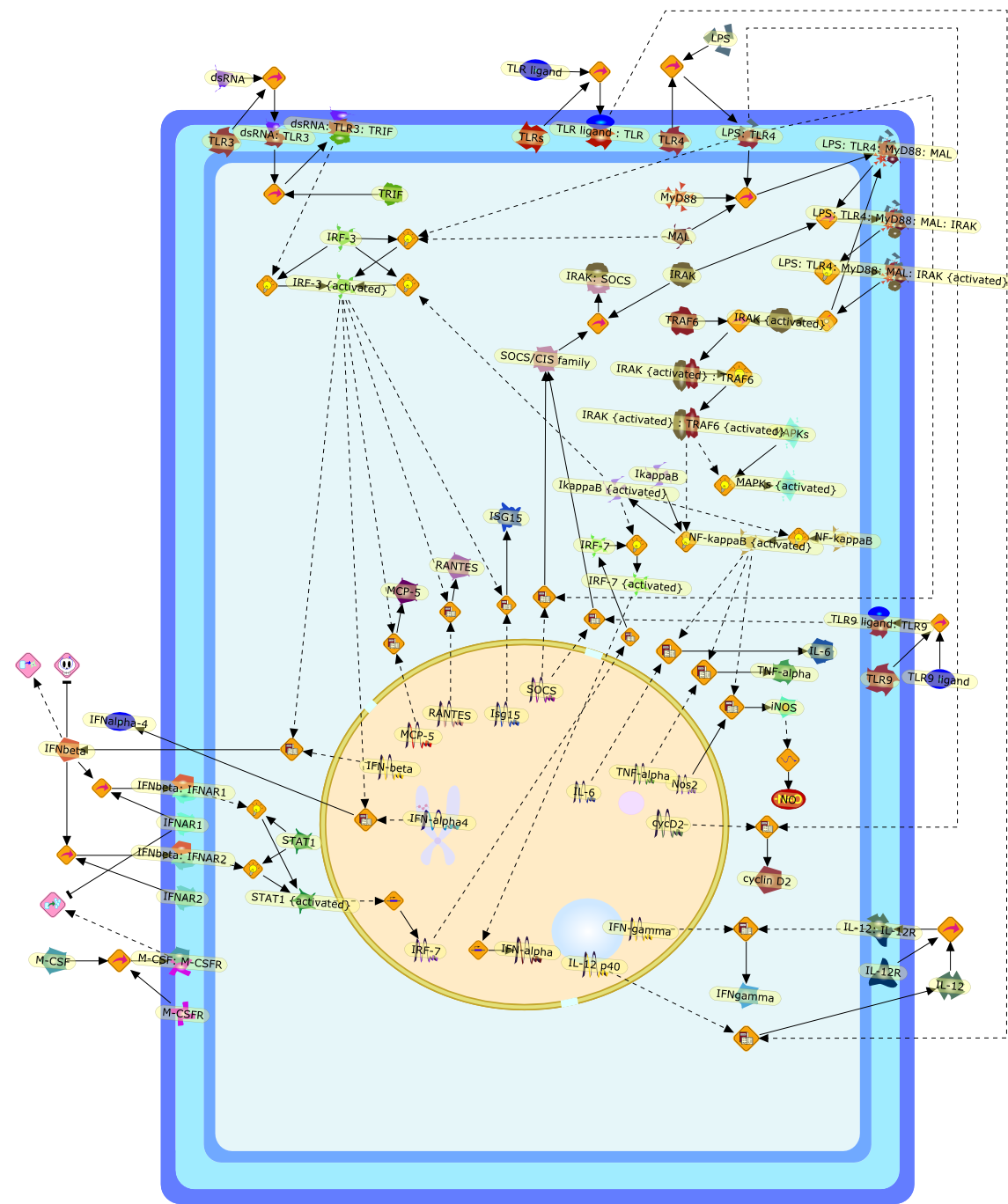
The interferon in TLR signaling: more than just antiviral.
Affiliation
Centre for Functional Genomics and Human Disease, Monash Institute ofReproduction and Development, Monash University, Clayton, Victoria 3168,Australia. paul.hertzog@med.monash.edu.au
Abstract
The Toll-like receptor (TLR) system is responsible for the recognition ofinfectious agents leading to initiation of the primary innate, and lateradaptive, immune response. Genetic technologies have enabled the discovery ofnew factors involved in these systems, their genetic manipulation and the globalanalyses of their effects on gene expression. Furthermore, this increasedunderstanding has resulted in the need to reassess our preconceptions about thefunctions of well-known molecules. For example, type I interferons (IFNs), whichwere discovered as antiviral proteins, are now known to be produced in responseto TLR activation by many pathogens, including bacteria. Should we be surprised?Has the inflammatory response unexpectedly highjacked the body's antiviralsystem? Or are we too easily blinkered by preconceptions from how a compound wasdiscovered?
PMID
14552837
|