| Original Literature | Model OverView |
|---|---|
|
Publication
Title
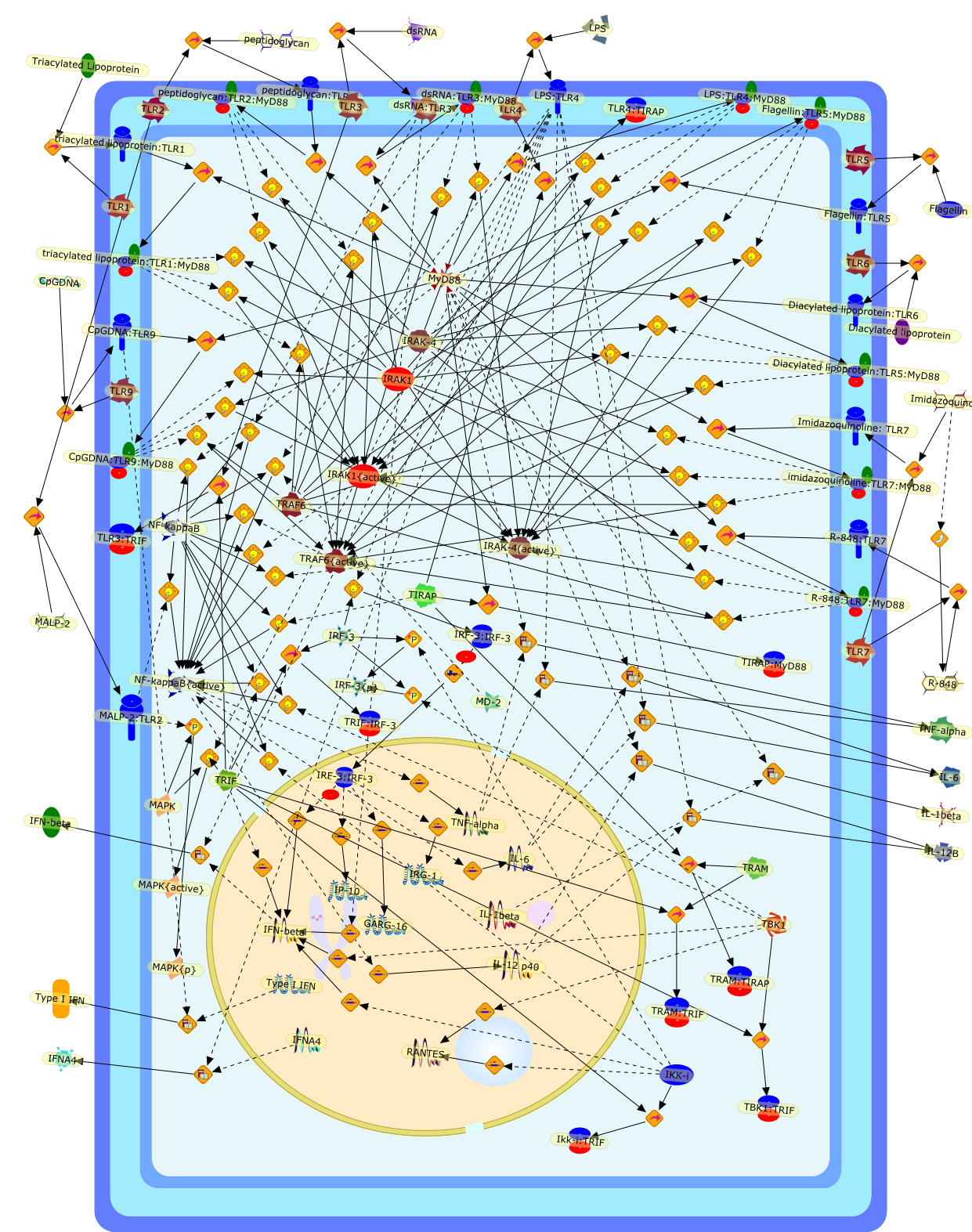
TIR domain-containing adaptors define the specificity of TLR signaling.
Affiliation
Department of Host Defense, Research Institute for Microbial Diseases, OsakaUniversity, 3-1 Yamada-oka, Suita, Osaka 565-0871, Japan.
Abstract
The concept that Toll-like receptors (TLRs) recognize specific molecularpatterns in various pathogens has been established. In signal transduction viaTLRs, MyD88, which harbors a Toll/IL-1 receptor (TIR)-domain and a death domain,has been shown to link between TLRs and MyD88-dependent downstream eventsleading to proinflammatory cytokine production and splenocyte proliferation.However, recent studies using MyD88-deficient mice have revealed that some TLRspossess a MyD88-independent pathway, which is represented by interferon(IFN)-beta production induced by LPS stimulation. This indicates that additionalsignaling molecules other than MyD88 exist in the TLR signaling pathway. Indeed,two additional TIR domain-containing adaptors, TIRAP/Mal and TRIF, have recentlybeen identified. Both define the specific biological responses of each TLR.
PMID
14698224
|