| Original Literature | Model OverView |
|---|---|
|
Publication
Title
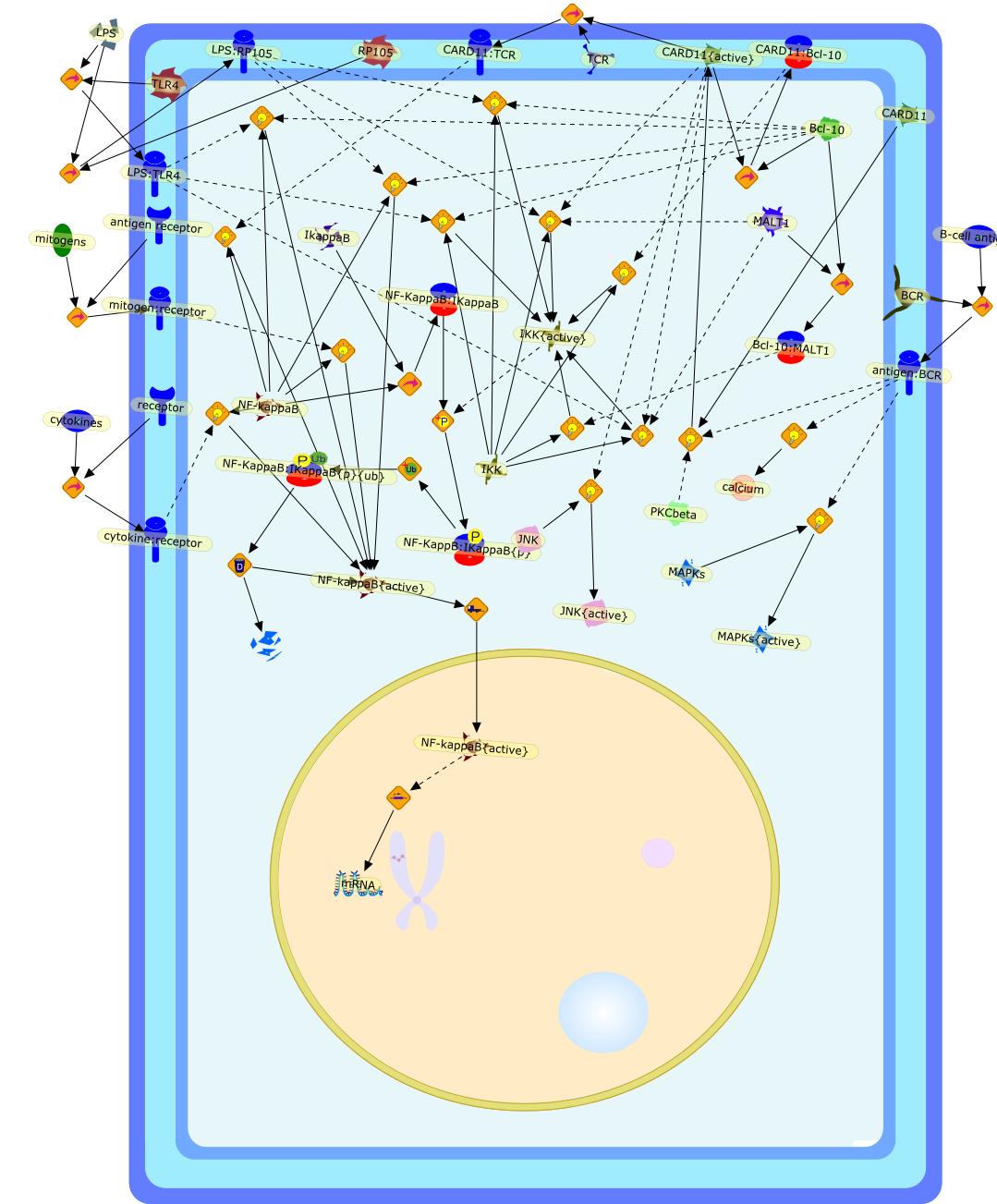
New roles for Bcl10 in B-cell development and LPS response.
Affiliation
Immune Receptor Signaling Group, Department of Physiological Chemistry,University of Ulm, Albert-Einstein-Allee 11, 89081 Ulm, Germany.klaus.fischer@medizin.uni-ulm.de
Abstract
PMID
15049289
|