| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Post-transcriptional regulation of proinflammatory proteins.
Affiliation
Immunology and Allergy, Brigham and Women's Hospital, Smith 652, One Jimmy FundWay, Boston, MA 02115, USA. panderson@rics.bwh.harvard.edu
Abstract
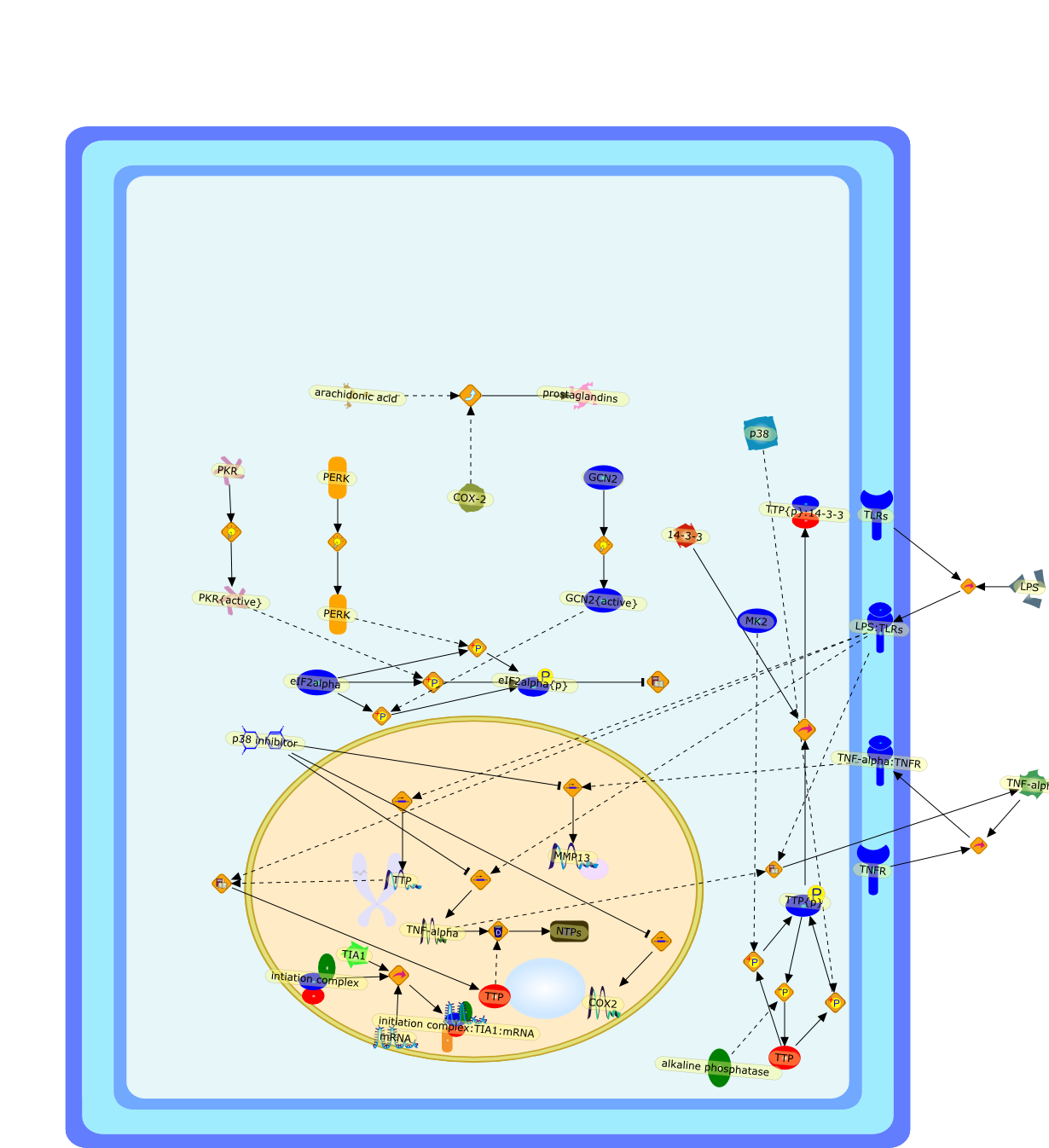
Post-transcriptional mechanisms play a critical role in regulating theexpression of numerous proteins that promote inflammatory arthritis. The mRNAsencoding a subset of these proteins possess adenine/uridine-rich elements (AREs)in their 3'-untranslated regions that profoundly influence the rate at whichmRNA is degraded and translated into protein. Tristetraprolin (TTP) and T cellintracellular antigen-1 (TIA-1) are ARE-binding proteins that dampen theexpression of this class of proteins by promoting mRNA degradation and proteintranslation, respectively. We have discovered that TIA-1 and TTP function asarthritis-suppressor genes: TIA-1-/- mice develop mild arthritis, TTP-/- micedevelop severe arthritis, and TIA-1-/-TTP-/- mice develop very severe arthritis.Paradoxically, lipopolysaccharide (LPS)-activated macrophages derived fromTIA-1-/-TTP-/- macrophages produce less tumor necrosis factor alpha (TNF-alpha)than TIA-1-/- or TTP-/- macrophages. The bone marrows of these mice exhibitincreased cellularity, reflecting the presence of mature neutrophils thatsecrete TNF-alpha in response to LPS stimulation. We hypothesize thatTIA-1-/-TTP-/- neutrophils are a source of arthritigenic TNF-alpha, whichpromotes severe erosive arthritis in these mice.
PMID
15075353
|