| Original Literature | Model OverView |
|---|---|
|
Publication
Title
A Toll-like receptor in horseshoe crabs.
Affiliation
Department of Biology, Faculty of Sciences, Kyushu University, Fukuoka, Japan.
Abstract
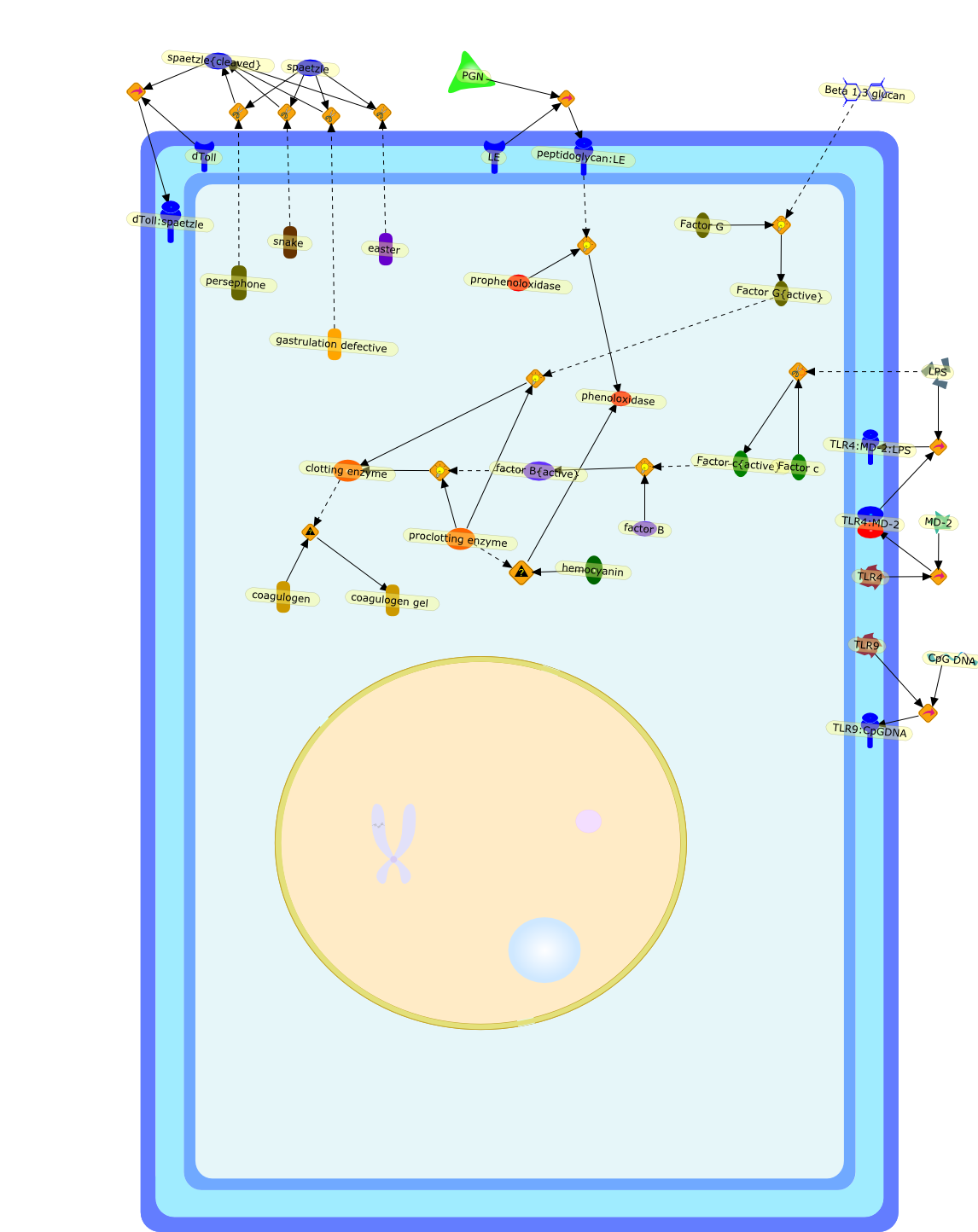
Non-self-recognition of invading microbes relies on the pattern-recognition ofpathogen-associated molecular patterns (PAMPs) derived from microbial cell-wallcomponents. Insects and mammals conserve a signaling pathway of the innateimmune system through cell-surface receptors called Tolls and Toll-likereceptors (TLRs). Bacterial lipopolysaccharides (LPSs) are an important triggerof the horseshoe crab's innate immunity to infectious microorganisms. Horseshoecrabs' granular hemocytes respond specifically to LPS stimulation, inducing thesecretion of various defense molecules from the granular hemocytes. Here, weshow a cDNA which we named tToll, coding for a TLR identified from hemocytes ofthe horseshoe crab Tachypleus tridentatus. tToll is most closely related toDrosophila Toll in both domain architecture and overall length. Human TLRs havebeen suggested to contain numerous PAMP-binding insertions located in theleucine-rich repeats (LRRs) of their ectodomains. However, the LRRs of tTollcontained no obvious PAMP-binding insertions. Furthermore, tToll wasnon-specifically expressed in horseshoe crab tissues. These observations suggestthat tToll does not function as an LPS receptor on granular hemocytes.
PMID
15199958
|