| Original Literature | Model OverView |
|---|---|
|
Publication
Title
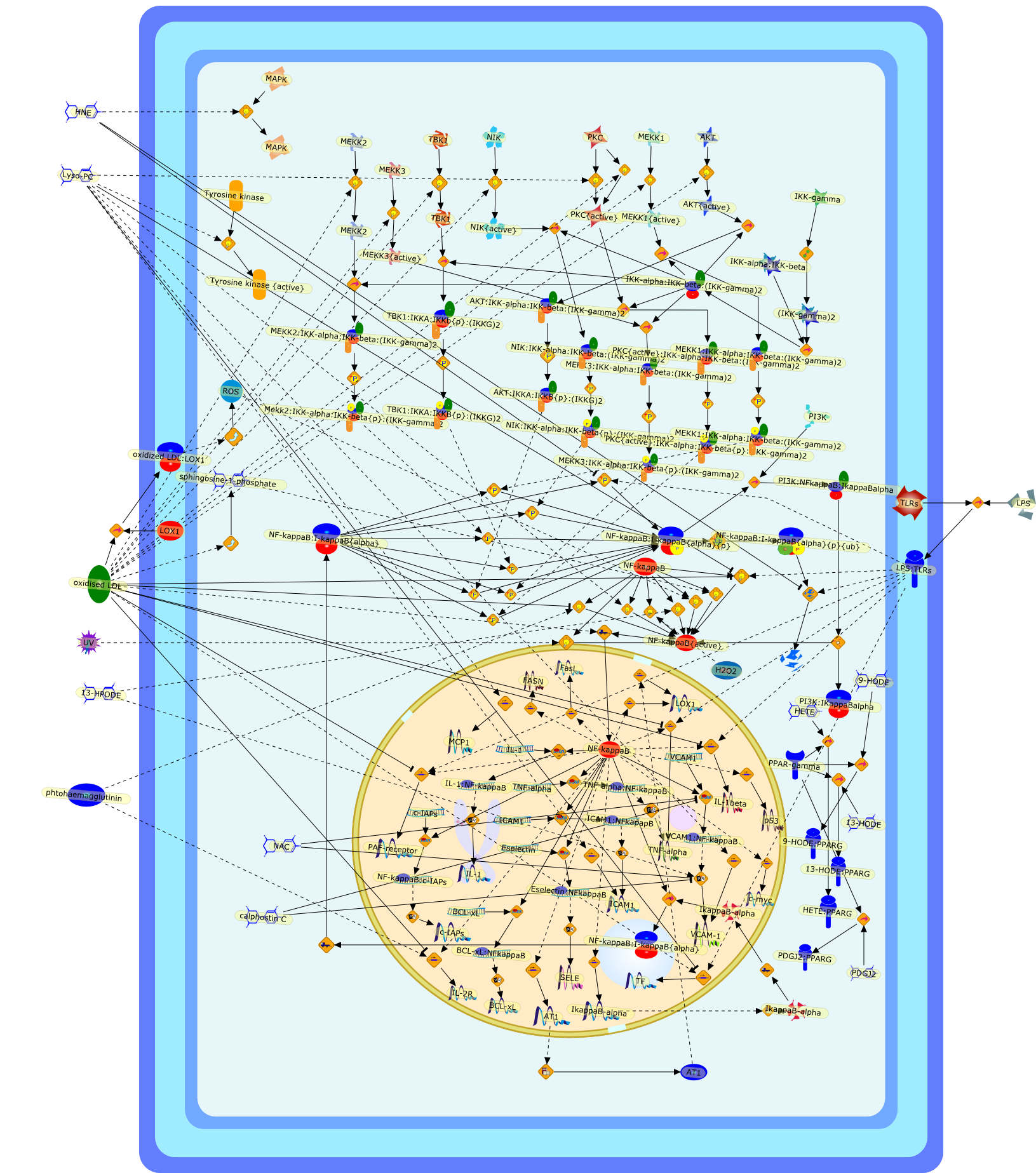
Dual role of oxidized LDL on the NF-kappaB signaling pathway.
Affiliation
INSERM U-466, CHU Rangueil, 31403 Toulouse, Cedex 4, France.
Abstract
Atherosclerosis is a slowly evolutive age-linked disease of large arteries,characterized by a local lipid deposition associated with a chronic inflammatoryresponse, leading potentially to acute plaque rupture, thrombosis and ischemicheart disease. Atherogenesis is a complex sequence of events associating firstexpression of adhesion molecules, recruitment of mononuclear cells to theendothelium, local activation of leukocytes and inflammation, lipid accumulationand foam cell formation. Low density lipoproteins (LDLs) become atherogenicafter undergoing oxidation by vascular cells, that transform them into highlybioreactive oxidized LDL (oxidized LDLs). Oxidized LDLs are involved in foamcell formation, and trigger proatherogenic events such as overexpression ofadhesion molecules, chemoattractant agents growth factors and cytokines involvedin the inflammatory process, cell proliferation and apoptosis. Moreover, thistoxic effect of oxidized LDLs plays probably a role in plaque erosion/ruptureand subsequent atherothrombosis. Several biological effects of oxidized LDLs aremediated through changes in the activity of transcription factors andsubsequently in gene expression. Oxidized LDLs exert a biphasic effect on theredox-sensitive transcription factor NF-kappaB, which can be activated therebyup-regulating proinflammatory gene expression, such as adhesion molecules,tissue factor, scavenger receptor LOX-1. On the other hand, higherconcentrations of oxidized LDLs may inhibit NF-kappaB activation triggered byinflammatory agents such as LPS, and may thereby exert an immunosuppressiveeffect. This review is an attempt to clarify the mechanism by which oxidizedLDLs may up- or down-regulate NF-kappaB, the role of NF-kappaB activation (orinhibition), and the consequences of the oxidized LDLs-mediated NF-kappaBdysregulation and their potential involvement in atherosclerosis.
PMID
15346645
|