| Original Literature | Model OverView |
|---|---|
|
Publication
Title
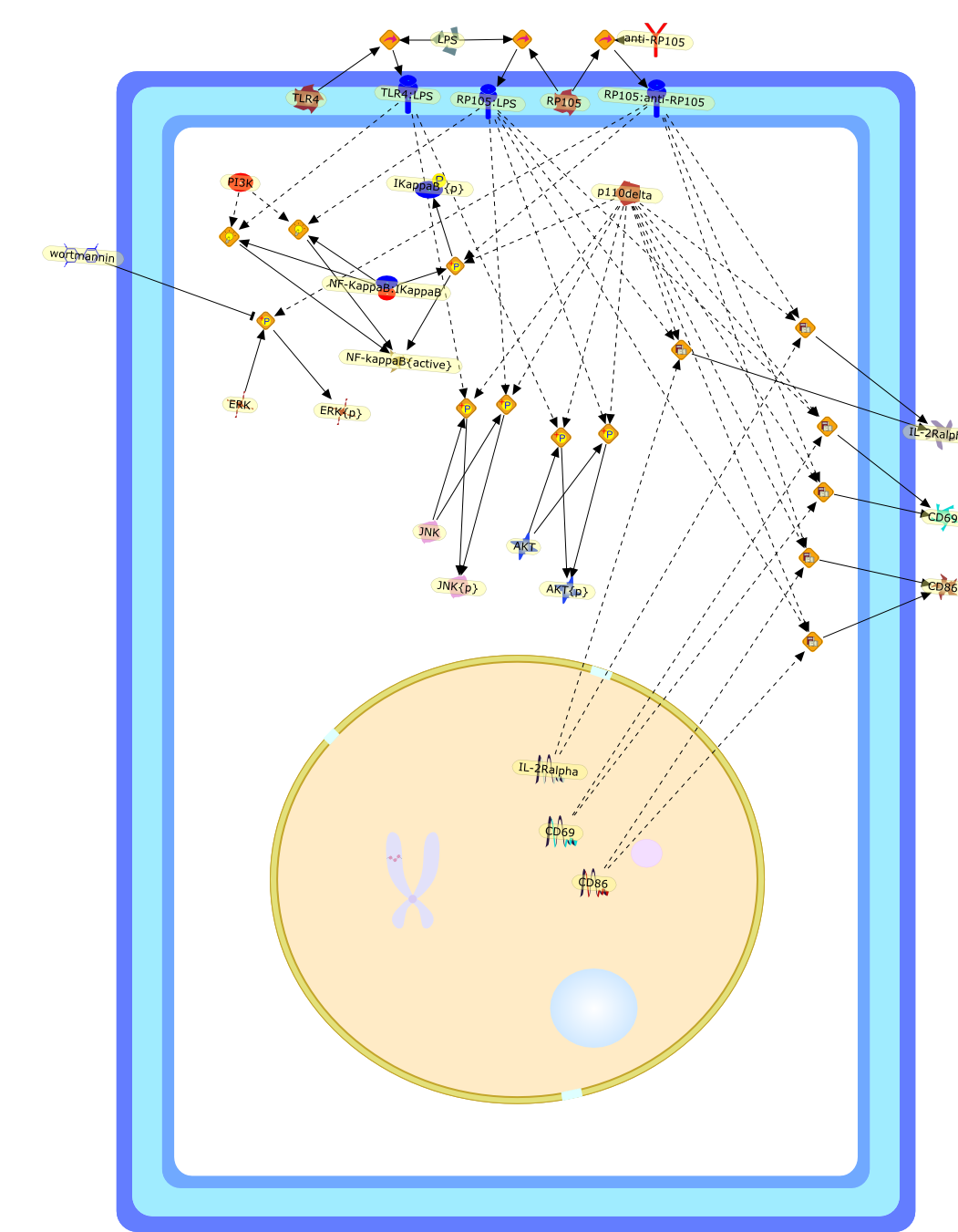
The p110delta subunit of phosphoinositide 3-kinase is required for thelipopolysaccharide response of mouse B cells.
Affiliation
Laboratory of Lymphocyte Signalling and Development, Babraham Institute,Babraham Research Campus, Cambridge CB2 4AT, UK. barbara.j.hebeis@gsk.com
Abstract
PI3K (phosphoinositide 3-kinase) I(A) family members contain a regulatorysubunit and a catalytic subunit. The p110delta catalytic subunit is expressedpredominantly in haematopoietic cells. There, among other functions, itregulates antigen receptor-mediated responses. Using mice deficient in thep110delta subunit of PI3K, we investigated the role of this subunit in LPS(lipopolysaccharide)-induced B cell responses, which are mediated by Toll-likereceptor 4 and RP105. After injection of DNP-LPS (where DNP stands for2,4-dinitrophenol), p110delta(-/-) mice produced reduced levels of DNP-specificIgM and IgG when compared with wild-type mice. In vitro, the proliferation andup-regulation of surface activation markers such as CD86 and CD25 induced by LPSand an antibody against RP105 were decreased. We analysed the activation stateof key components of the LPS pathway in B cells to determine whether there was adefect in signalling in p110delta(-/-) B cells. They showed normalextracellular-signal-regulated kinase phosphorylation, but anti-RP105-inducedprotein kinase B, IkappaB (inhibitor of nuclear factor kappaB) and c-JunN-terminal kinase activation was severely reduced. This demonstrates that thep110delta subunit of PI3K is involved in the LPS response in B cells and mayrepresent a link between the innate and the adaptive immune system.
PMID
15494016
|