| Original Literature | Model OverView |
|---|---|
|
Publication
Title
The SAP family of adaptors in immune regulation.
Affiliation
Unite INSERM U429, Hopital Necker Enfants-Malades, 149 rue de Sevres, 75015Paris, France. latour@necker.fr
Abstract
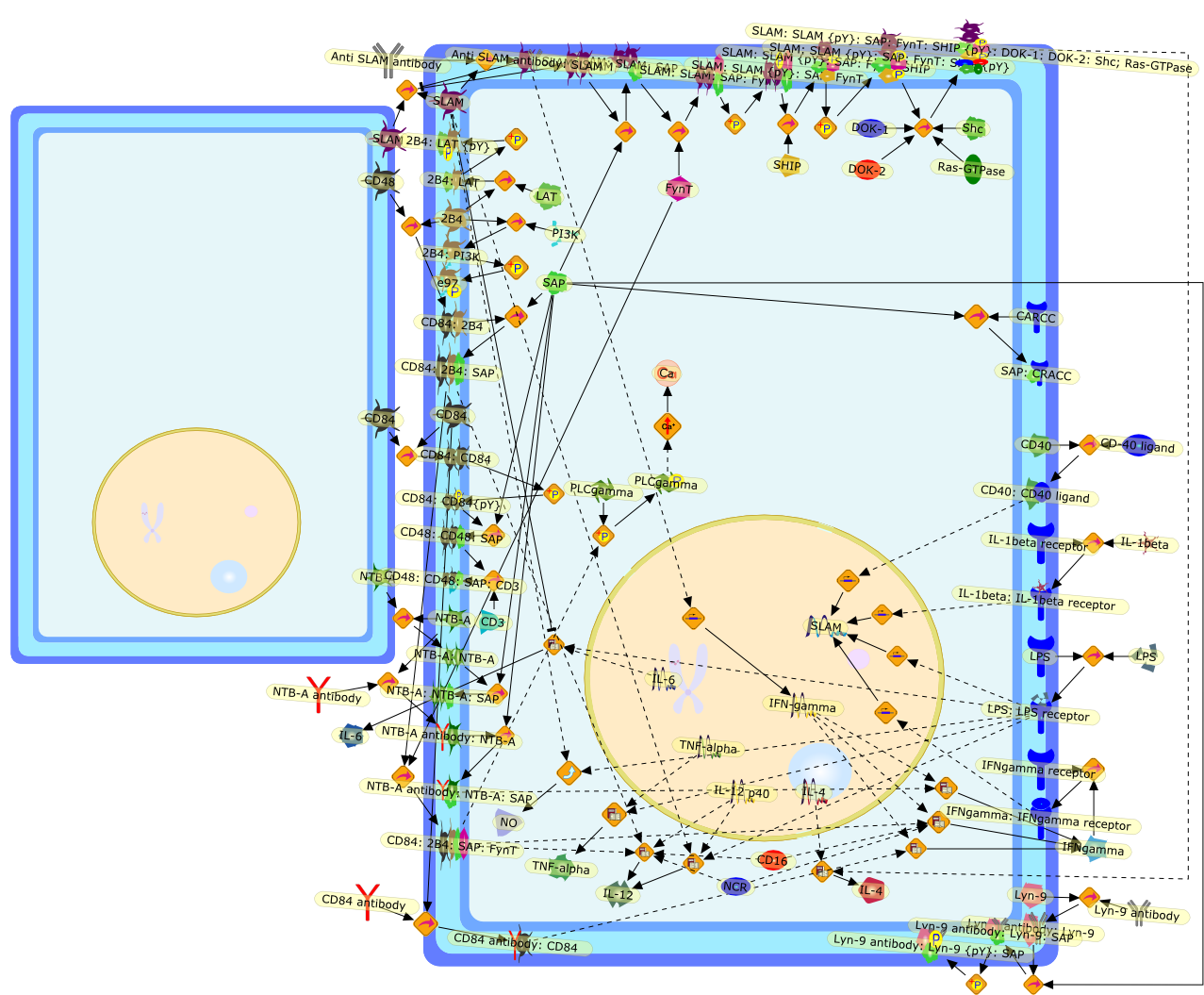
SAP and EAT-2 define a new class of adaptor proteins composed almost exclusivelyof a Src homology 2 (SH2) domain. By way of their SH2 domain, SAP-like adaptorsinteract with tyrosine-based motifs in the cytoplasmic region of SLAM-relatedreceptors, a family of immune cell-specific molecules involved inimmunoregulation. Recent findings indicate that SAP is required for thefunctions of SLAM family receptors, as a consequence of its ability to promoterecruitment of Src-related protein tyrosine kinase FynT and allow SLAM-relatedreceptors to transduce tyrosine phosphorylation signals. SAP is mutated inX-linked lymphoproliferative (XLP) syndrome, a rare inherited human diseasecharacterized by an deregulated immune response to Epstein-Barr virus infection.Several lines of evidence indicate that defects in the activities ofSLAM-related receptors caused by SAP deficiency account for the immunedysfunctions associated with XLP.
PMID
15541655
|