| Original Literature | Model OverView |
|---|---|
|
Publication
Title
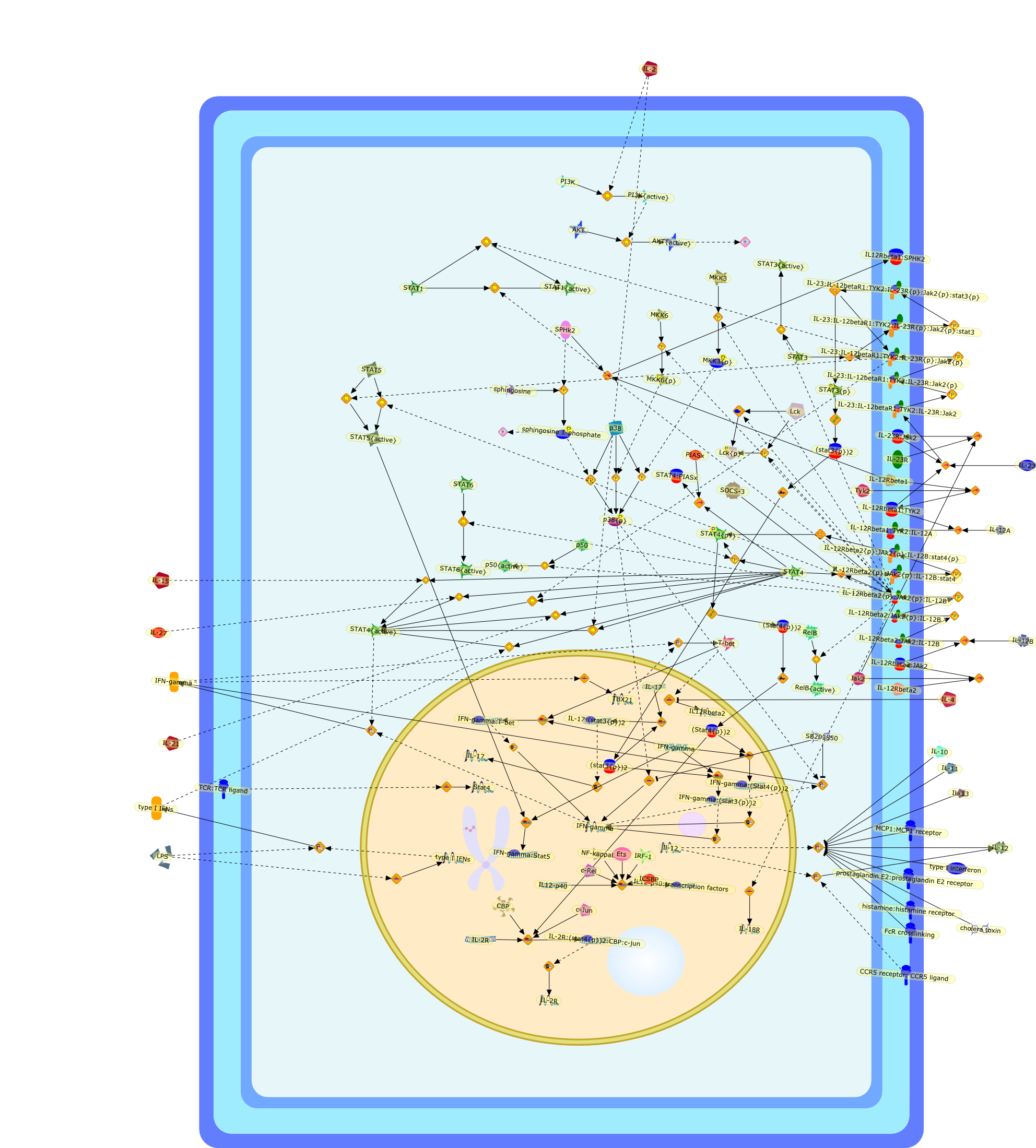
Signaling by IL-12 and IL-23 and the immunoregulatory roles of STAT4.
Affiliation
Molecular Immunology & Inflammation Branch, NIAMS, National Institutes ofHealth, Bethesda, MD 20892-1820, USA.
Abstract
Produced in response to a variety of pathogenic organisms, interleukin (IL)-12and IL-23 are key immunoregulatory cytokines that coordinate innate and adaptiveimmune responses. These dimeric cytokines share a subunit, designated p40, andbind to a common receptor chain, IL-12R beta 1. The receptor for IL-12 iscomposed of IL-12R beta 1 and IL-12R beta 2, whereas IL-23 binds to a receptorcomposed of IL-12R beta 1 and IL-23R. Both cytokines activate the Janus kinasesTyk2 and Jak2, the transcription factor signal transducer and activator oftranscription 4 (STAT4), as well as other STATs. A major action of IL-12 is topromote the differentiation of naive CD4+ T cells into T-helper (Th) 1 cells,which produce interferon (IFN)-gamma, and deficiency of IL-12, IL-12R subunitsor STAT4 is similar in many respects. In contrast, IL-23 promotes end-stageinflammation. Targeting IL-12, IL-23, and their downstream signaling elementswould therefore be logical strategies for the treatment of immune-mediateddiseases.
PMID
15546391
|