| Original Literature | Model OverView |
|---|---|
|
Publication
Title
A novel negative regulator for IL-1 receptor and Toll-like receptor 4.
Affiliation
Division of Immunology, Infection and Inflammation, University of Glasgow,Glasgow G11 6NT, UK. fyl1h@clinmed.gla.ac.uk
Abstract
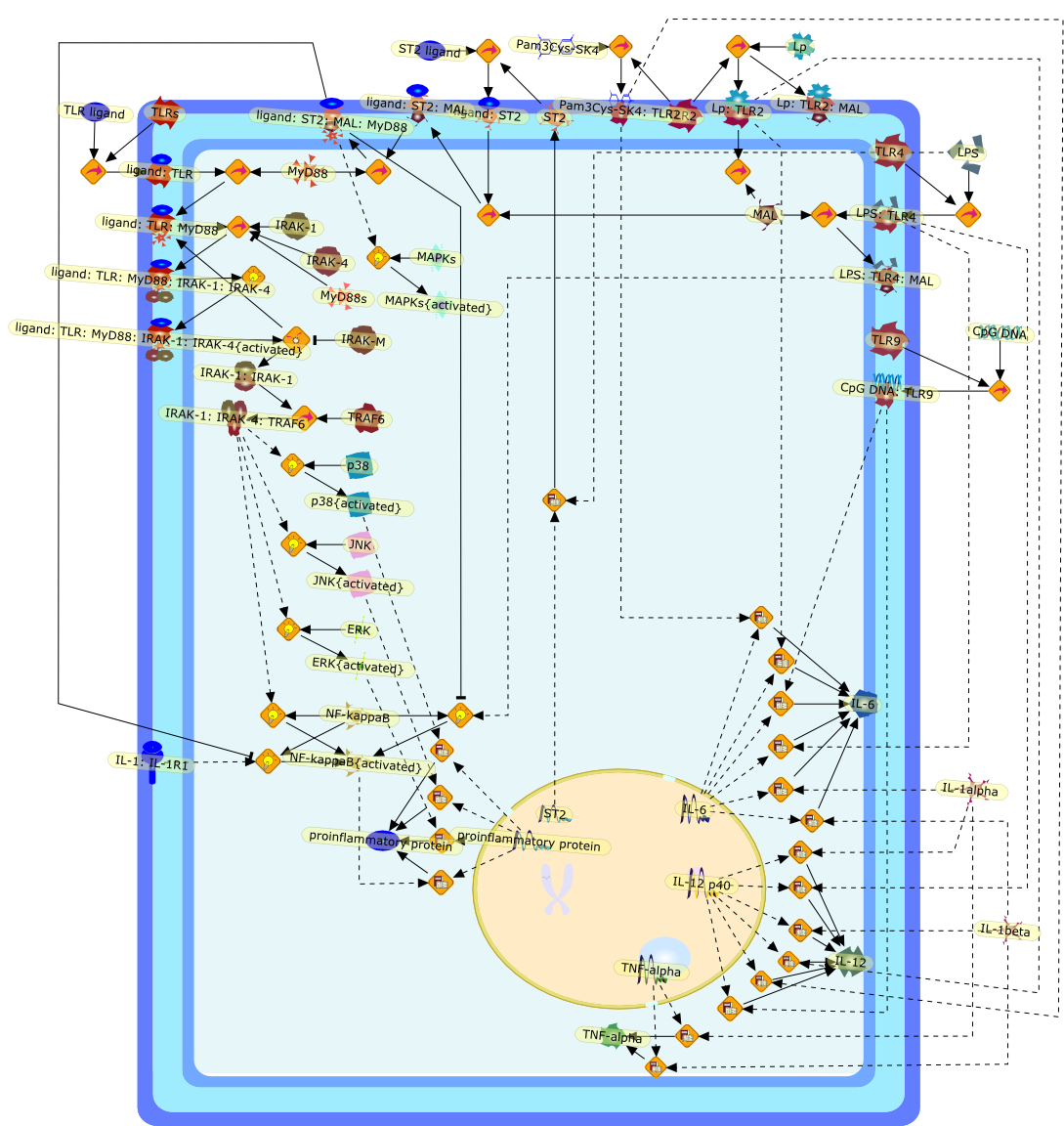
The Toll-IL-1 receptor (TIR) superfamily, defined by the presence of anintracellular TIR domain, initiates innate immunity via NF-kappaB activation,leading to production of proinflammatory cytokines. ST2 is a member of the TIRfamily that does not activate NF-kappaB and has been suggested as an importanteffector molecule of type 2 T helper cell responses. We have recentlydemonstrated that the membrane bound form of ST2 (ST2L) negatively regulatedIL-1RI and TLR4 but not TLR3 signaling by sequestrating the adaptors MyD88 andMal. In contrast to wild-type mice, ST2 deficient mice failed to developendotoxin tolerance. Thus, ST2 suppresses IL-1R and TLR4 signaling via MyD88-and Mal-dependent pathways and modulates innate immunity. The results provide amolecular explanation for the role of ST2 in T(H)2 responses since inhibition ofTLRs will promote a T(H)2 response and also identify ST2 as a key regulator ofendotoxin tolerance.
PMID
15585304
|