| Original Literature | Model OverView |
|---|---|
|
Publication
Title
TICAM-1 and TICAM-2: toll-like receptor adapters that participate in inductionof type 1 interferons.
Affiliation
Department of Immunology, Osaka Medical Center for Cancer and CardiovascularDiseases, Higashinari-ku, Osaka 537-8511, Japan. seya-tu@med.hokudai.ac.jp
Abstract
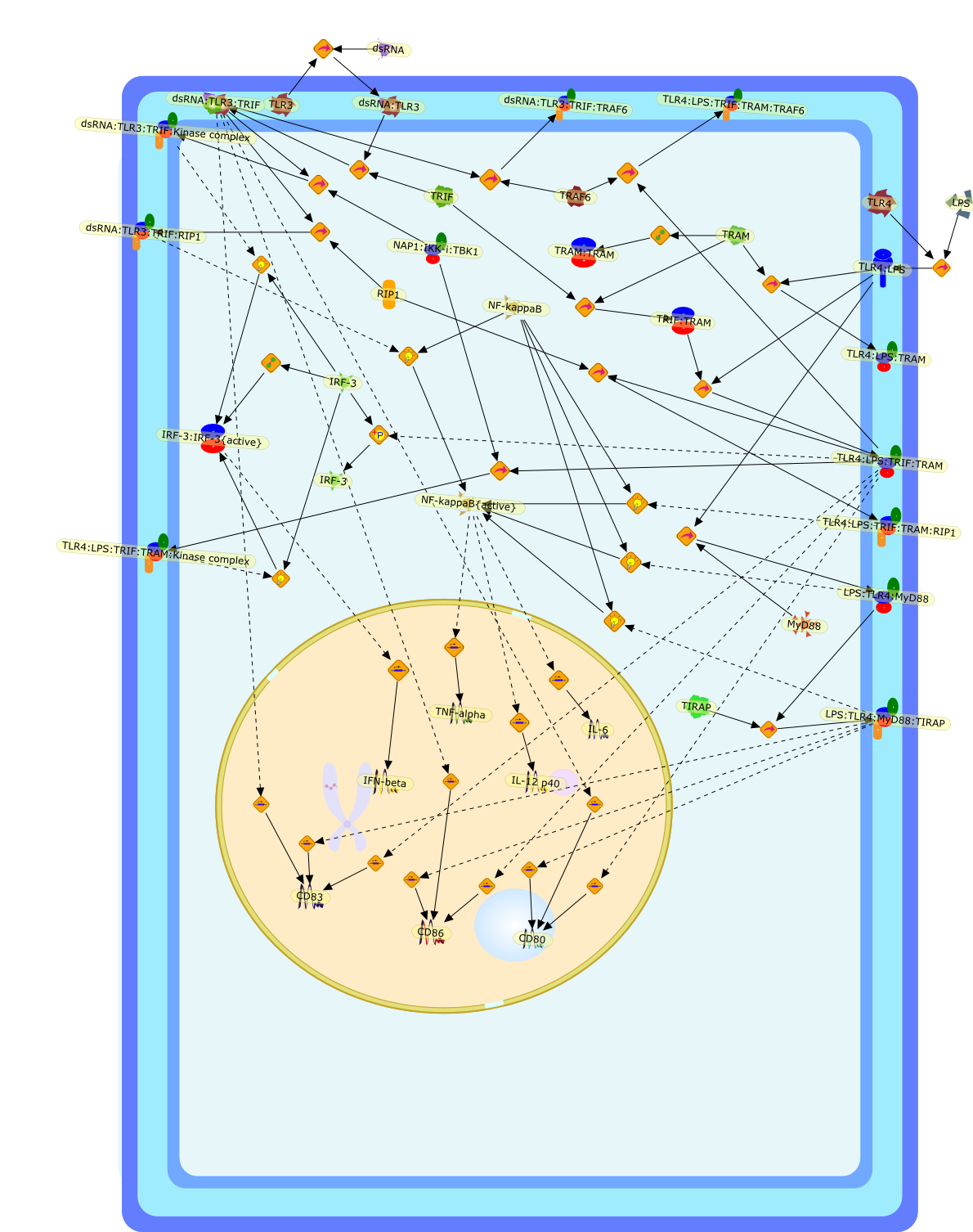
Toll-like receptor (TLR) 3 and 4 mediate the expression of many genes, includingNF-kappaB- and interferon-regulatory factor (IRF)-3/interferon (IFN)-induciblegenes, in macrophages and dendritic cells (DCs) in response to their ligandstimuli, polyI:C and lipopolysaccharide (LPS). Toll-IL-1 receptor homologydomain (TIR)-containing adapter molecule 1 (TICAM-1) facilitates expression ofIFN-inducible genes via TLR3. Although MyD88 and Mal/TIRAP adapters functiondownstream of TLR4, they barely induce IFN-beta. In addition, DC maturation aswell as IFN-beta induction are largely independent of MyD88 and Mal/TIRAP.TICAM-1 is the functional adapter for both TLR3 and TLR4 that induces type 1 IFNand MyD88-independent DC maturation. In LPS-mediated TLR4 activation, a complexof TICAM-1 and an additional TLR4-binding adapter serves as the adapter. Wenamed this TLR4-TICAM-1-bridging adapter TICAM-2. Our results reveal the detailsof MyD88-independent pathways which separately recruit the distinct adaptersdownstream of TLR3 and TLR4 and variations of the TLR output are in partregulated by the two additional adapters in DCs.
PMID
15618008
|