| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Modulation of Toll-interleukin 1 receptor mediated signaling.
Affiliation
Department of Immunology, Lerner Research Institute, Cleveland ClinicFoundation, 9500 Euclid Avenue, Cleveland, OH 44195, USA. lix@ccf.org
Abstract
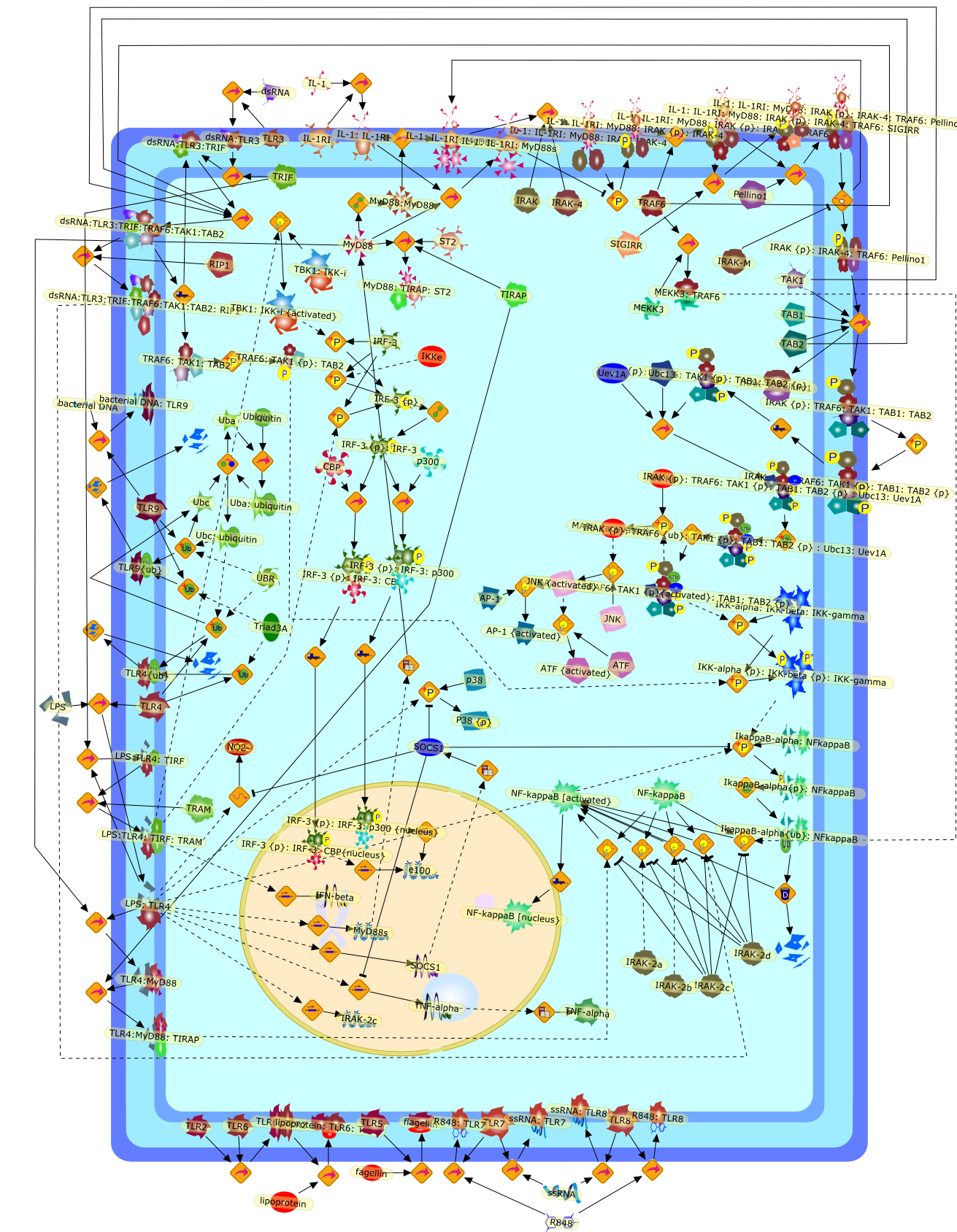
Toll-like receptors (TLRs) belong to the Toll-interleukin 1 receptorsuperfamily, which is defined by a common intracellular Toll-IL-1 receptor (TIR)domain. A group of TIR domain containing adaptors (MyD88, TIRAP, TRIF and TRAM),are differentially recruited to the Toll-IL-1 receptors, contributing to thespecificity of signaling. The IL-1 mediated signaling pathway serves as a"prototype" for other family members. Genetic and biochemical studies revealthat IL-1R uses adaptor molecule MyD88 to mediate a very complex pathway,involving a cascade of kinases organized by multiple adapter molecules intosignaling complexes, leading to activation of the transcription factor NFkappaB.Several Toll-like receptors utilize variations of the "prototype" pathway byemploying different adaptor molecules. Double-stranded RNA triggered,TLR3-mediated signaling is independent of MyD88, IRAK4, and IRAK. The adaptermolecule TRIF is utilized by TLR3 to mediate the activation of NFkappaB andIRF3. LPS-induced, TLR4-mediated signaling employs multiple TIR-domaincontaining adaptors, MyD88/TIRAP to mediate NFkappaB activation, TRIF/TRAM forIRF3 activation. Recent studies have also begun to unravel how these pathwaysare negatively regulated. SIGIRR (also known as TIR8), a member of TIRsuperfamily that does not activate the transcription factors NFkappaB and IRF3,instead negatively modulates responses. Cells from SIGIRR-null mice showenhanced activation in response to either IL-1 or certain Toll ligands. Inaddition to SIGIRR, several other negative regulators have been shown to inhibitthe TIR signaling, including ST2, IRAKM, MyD88s, SOCS1, and Triad3A. Thecoordinated positive and negative regulation of the TIR signaling ensures theappropriate modulation of the innate and inflammatory responses.
PMID
15662540
|