| Original Literature | Model OverView |
|---|---|
|
Publication
Title
The serine/threonine kinase Pim-1.
Affiliation
Institut fur Zellbiologie (Tumorforschung), Universitaetsklinikum Essen, IFZ,Virchowstrasse 173, D-45122 Essen, Germany.
Abstract
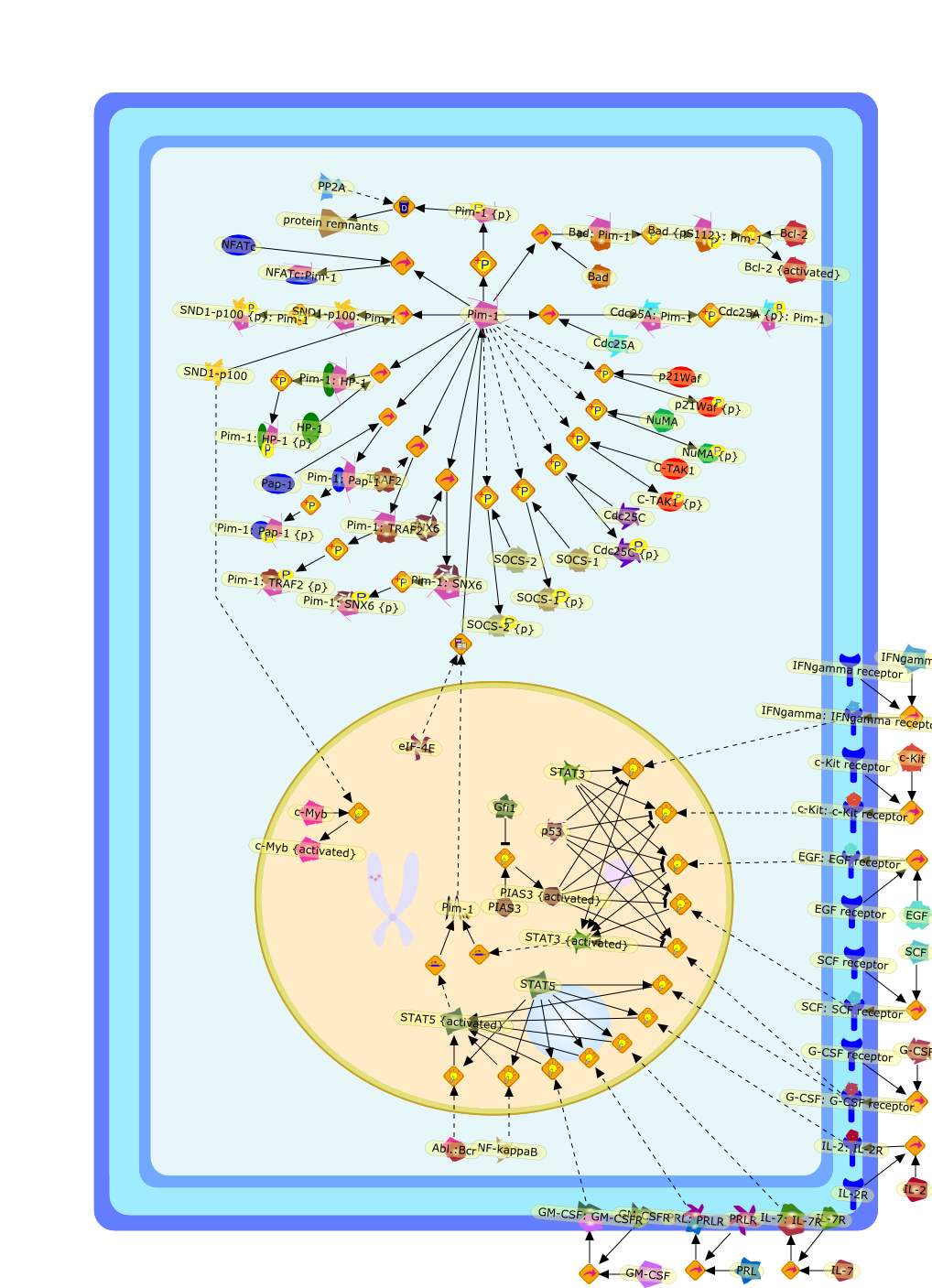
The human pim-1 gene encodes a serine/threonine kinase, which belongs to thegroup of calcium/calmodulin-regulated kinases (CAMK). It contains acharacteristic kinase domain, a so-called ATP anchor and an active site. Inmouse and human, two Pim-1 proteins are produced from the same gene by using analternative upstream CUG initiation codon, a 44 kD and another, shorter 34 kDform that both contain the kinase domain. Expression of Pim-1 is widespread andranges from the hematopoietic and lymphoid system to prostate, testis and oralepithelial cells. Two other proteins with significant sequence similaritiesexist, Pim-2 and Pim-3; both are also serine/threonine kinases and have largelyoverlapping functions. Pim-1 is able to phosphorylate different targets, most ofwhich are involved in cell cycle progression or apoptosis. Pim-1 expression canbe induced by several external stimuli in particular by a number of cytokinesrelevant in the immune system, which led to the labeling of Pim-1 as a "booster"for the immune response.
PMID
15694833
|