| Original Literature | Model OverView |
|---|---|
|
Publication
Title
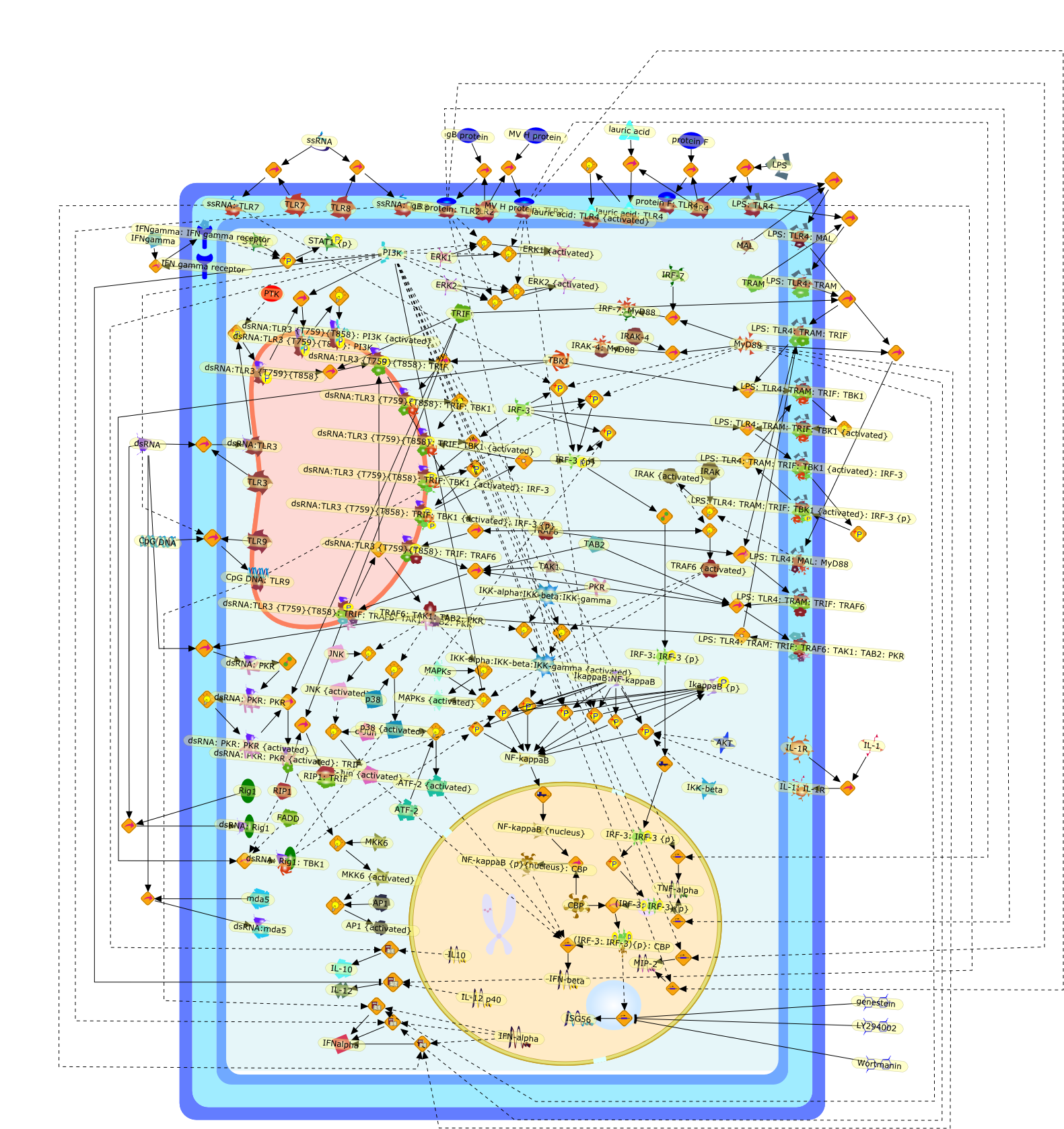
Transcriptional signaling by double-stranded RNA: role of TLR3.
Affiliation
Department of Molecular Genetics/NC20, The Lerner Research Institute, TheCleveland Clinic Foundation, 9500 Euclid Avenue, Cleveland, OH 44195, USA.seng@ccf.org
Abstract
Mammalian Toll-like receptors recognize components of invading microbes andtrigger the first line of innate immune response that is mediated bytranscriptional induction of a large number of cellular genes. Toll-likereceptor 3 (TLR3) is thought to be a major mediator of cellular response toviral infection, because it responds to double-stranded (ds) RNA, a commonby-product of viral replication. This article is focused on the nature of thesignaling pathways activated by TLR3 and dsRNA. The genes induced by TLR3activation include those that encode secreted antiviral cytokines, such asinterferon (IFN), and those that encode intracellular viral stress-inducibleproteins. Recent studies have revealed several unique features of TLR3 signalingthat are highlighted here. Specifically, we discuss the roles of receptortyrosine phosphorylation, PI3 kinase and two-step activation of thetranscription factors, IRF-3 and NF-kappaB, in mediating TLR3-signaling.
PMID
15733829
|