| Original Literature | Model OverView |
|---|---|
|
Publication
Title
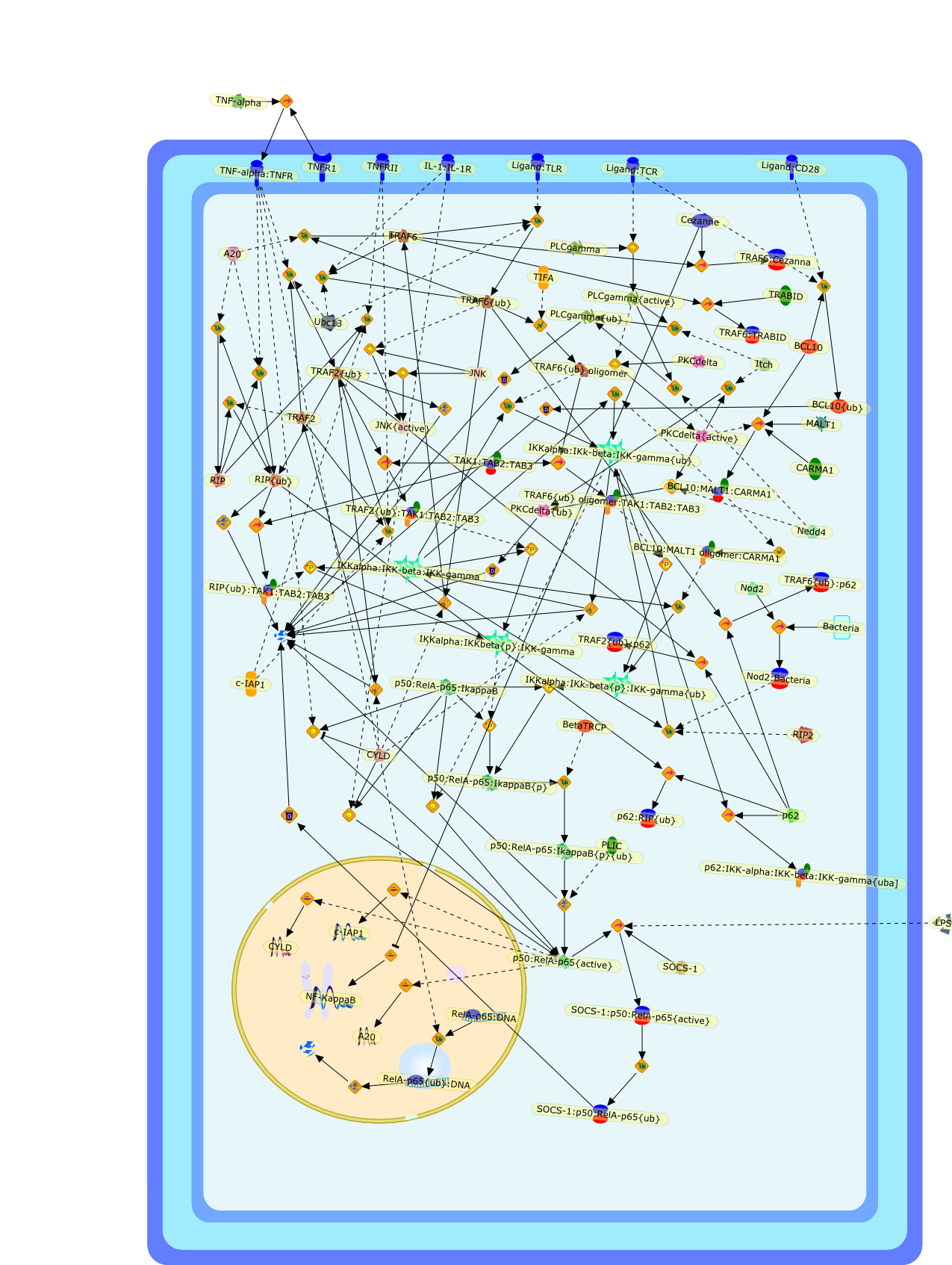
A pervasive role of ubiquitin conjugation in activation and termination ofIkappaB kinase pathways.
Affiliation
Max-Delbruck-Center for Molecular Medicine, Robert-Rossle-Strasse 10, D-13122Berlin, Germany.
Abstract
The nuclear factor (NF)-kappaB pathway is a paradigm for gene expression controlby ubiquitin-mediated protein degradation. In stimulated cells, phosphorylationby the IkappaB kinase (IKK) complex primes NF-kappaB-inhibiting IkappaBmolecules for lysine (Lys)-48-linked polyubiquitination and subsequentdestruction by the 26S proteasome. However, recent studies indicate that theubiquitin (Ub) system controls NF-kappaB pathways at many levels. Ub ligases areactivated by different upstream signalling pathways, and they function ascentral regulators of IKK and c-Jun amino-terminal kinase activation. Theassembly of Lys 63 polyUb chains provides docking surfaces for the recruitmentof IKK-activating complexes, a reaction that is counteracted by deubiquitinatingenzymes. Furthermore, Ub conjugation targets upstream signalling mediators aswell as nuclear NF-kappaB for post-inductive degradation to limit the durationof signalling.
PMID
15809659
|