| Original Literature | Model OverView |
|---|---|
|
Publication
Title
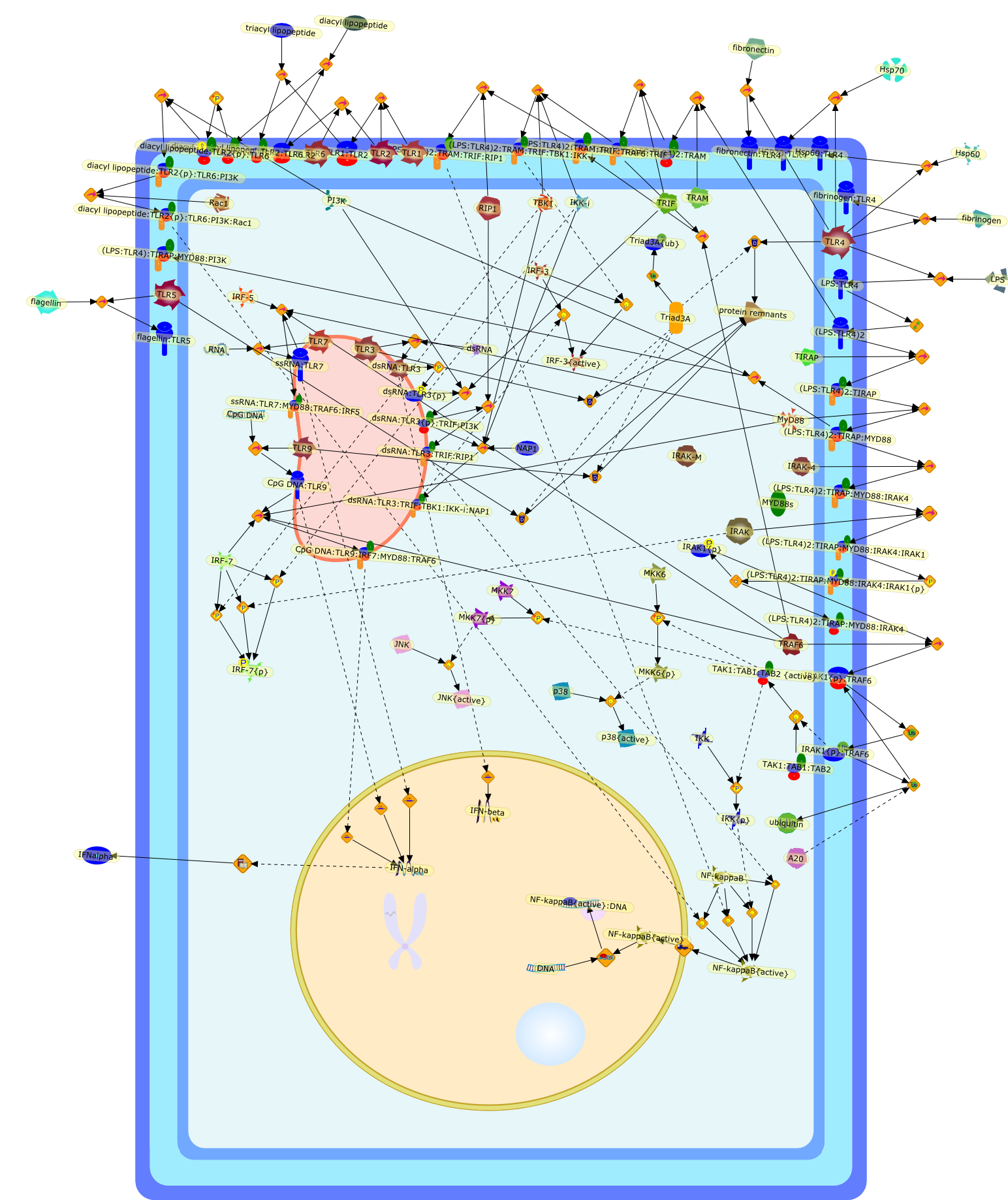
Adaptor usage and Toll-like receptor signaling specificity.
Affiliation
Department of Biochemistry, Trinity College Dublin, Dublin 2, Ireland.aidunne@tcd.ie
Abstract
It is now well established that Toll-like receptors (TLRs) act as primarysensors of microbial compounds. Details of the molecular mechanisms governingTLR responses are emerging steadily and our understanding of the signalingpathways activated these receptors has improved greatly over the last few years.Differences in adaptor usage, cellular localisation and signaling cascades havebeen elucidated. In this review we will summarize the current understanding ofTLR signaling and its regulation.
PMID
15876435
|