| Original Literature | Model OverView |
|---|---|
|
Publication
Title
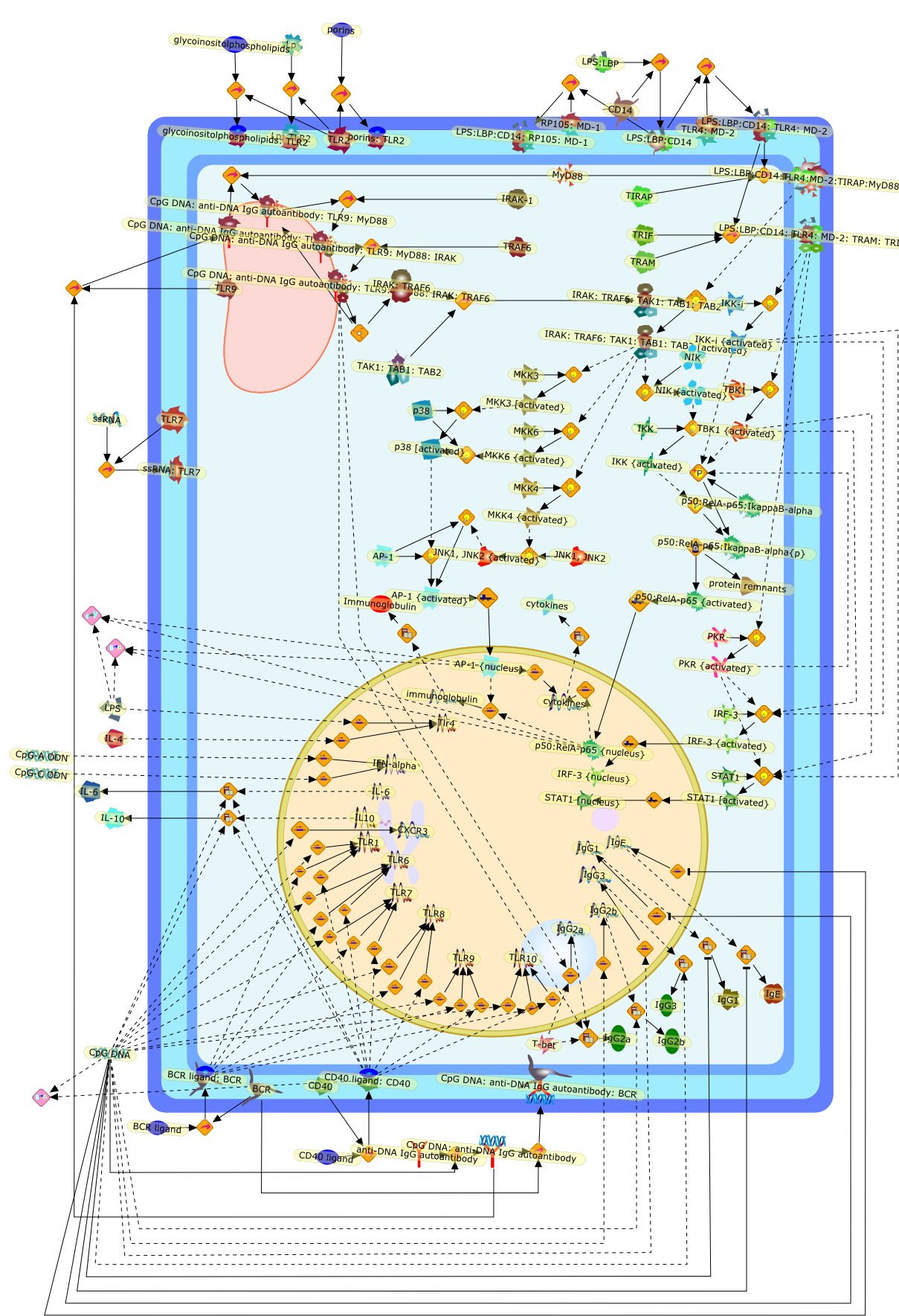
Signaling in B cells via Toll-like receptors.
Affiliation
Departments of Internal Medicine, Washington University School of Medicine, St.Louis, Missouri 63110, USA. speng@im.wustl.edu
Abstract
Toll-like receptors (TLRs) and their ligands have emerged as importantregulators of immunity, relevant to a wide range of effector responses fromvaccination to autoimmunity. The most well-studied ligands of TLRs expressed onB cells include the lipopolysaccharides (for TLR4) and CpG-containing DNAs (forTLR9), which induce and/or co-stimulate B cells to undergo proliferation, classswitching and differentiation into antibody-secreting cells. Recent developmentsin this area include advancements into our understanding of the role of thesereceptor pathways in B cells, and in particular the relevance of TLR9, which hasreceived substantial attention as both a Th1-like inflammatory immunomodulatorand a pathogenic co-stimulator of autoreactive B cell responses.
PMID
15886111
|