| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Cytokines, PGE2 and endotoxic fever: a re-assessment.
Affiliation
Department of Physiology, College of Medicine, The University of Tennessee,Health Science Center, Memphis, 38163, USA. blatteis@physio1.utmem.edu
Abstract
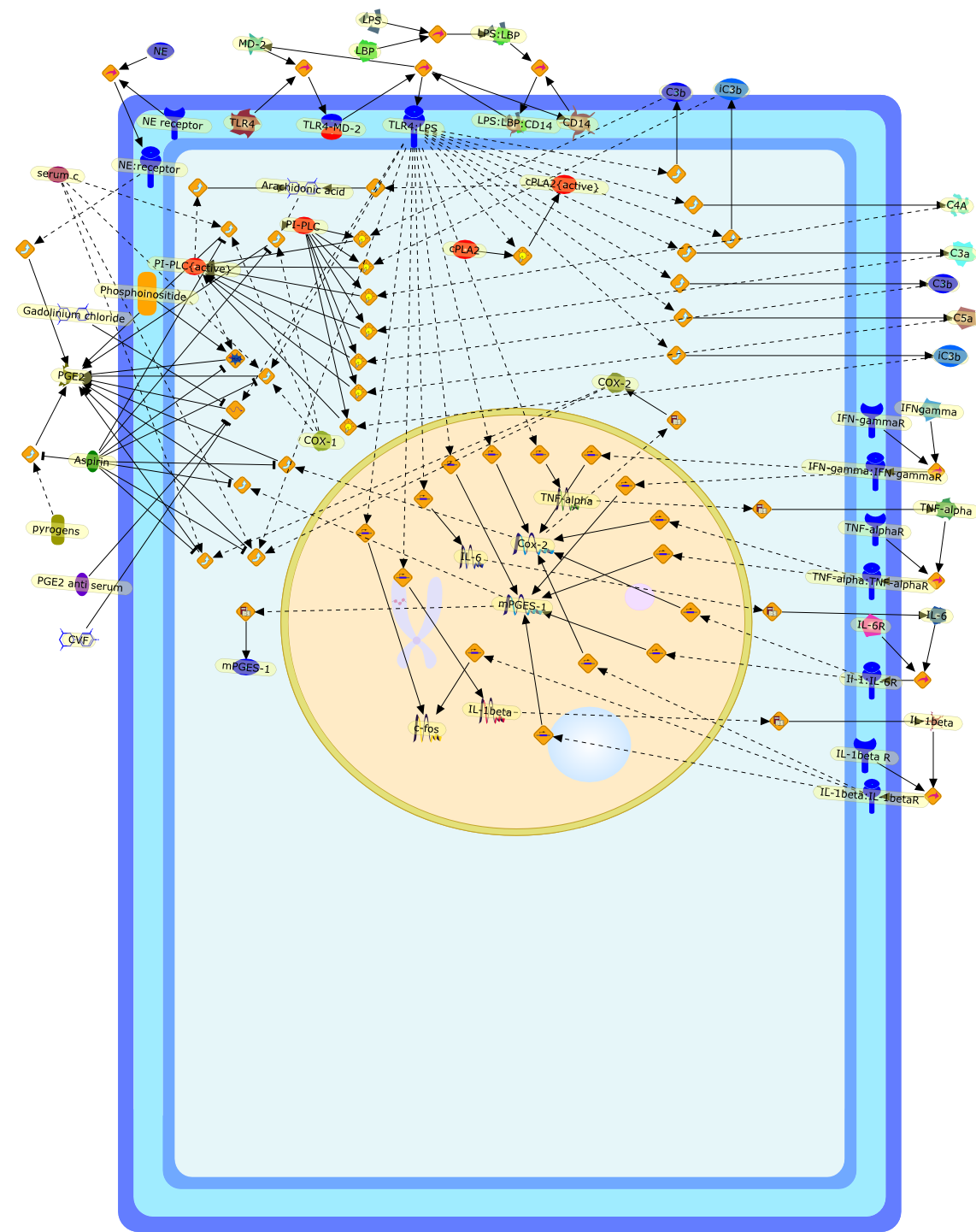
The innate immune system serves as the first line of host defense against thedeleterious effects of invading infectious pathogens. Fever is the hallmarkamong the defense mechanisms evoked by the entry into the body of suchpathogens. The conventional view of the steps that lead to fever production isthat they begin with the biosynthesis of pyrogenic cytokines by mononuclearphagocytes stimulated by the pathogens, their release into the circulation andtransport to the thermoregulatory center in the preoptic area (POA) of theanterior hypothalamus, and their induction there of cyclooxygenase(COX)-2-dependent prostaglandin (PG)E(2), the putative final mediator of thefebrile response. But data accumulated over the past 5 years have graduallychallenged this classical concept, due mostly to the temporal incompatibility ofthe newer findings with this concatenation of events. Thus, the former studiesgenerally overlooked that the production of cytokines and the transduction oftheir pyrogenic signals into fever-mediating PGE(2) proceed at relatively slowrates, significantly slower certainly than the onset latency of fever producedby the i.v. injection of bacterial endotoxic lipopolysaccharides (LPS). Here, wereview the conflicts between the earlier and the more recent findings andsummarize new data that reconcile many of the contradictions. A unified modelbased on these data explicating the generation and maintenance of the febrileresponse is presented. It postulates that the steps in the production of LPSfever occur in the following sequence: the immediate activation by LPS of thecomplement (C) cascade, the stimulation by the anaphylatoxic C component C5a ofKupffer cells, their consequent, virtually instantaneous release of PGE(2), itsexcitation of hepatic vagal afferents, their transmission of the induced signalsto the POA via the ventral noradrenergic bundle, and the activation by the thus,locally released norepinephrine (NE) of neural alpha(1)- and glialalpha(2)-adrenoceptors. The activation of the first causes an immediate,PGE(2)-independent rise in core temperature (T(c)) [the early phase of fever; anantioxidant-sensitive PGE(2) rise, however, accompanies this first phase], andof the second a delayed, PGE(2)-dependent T(c) rise [the late phase of fever].Meanwhile-generated pyrogenic cytokines and their consequent upregulation ofblood-brain barrier cells COX-2 also contribute to the latter rise. Theconsecutive steps that initiate the febrile response to LPS would now appear,therefore, to occur in an order different than conceived originally.
PMID
15967158
|