| Original Literature | Model OverView |
|---|---|
|
Publication
Title
The Toll-like receptors: analysis by forward genetic methods.
Affiliation
The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA,92037, USA. Bruce@scripps.edu
Abstract
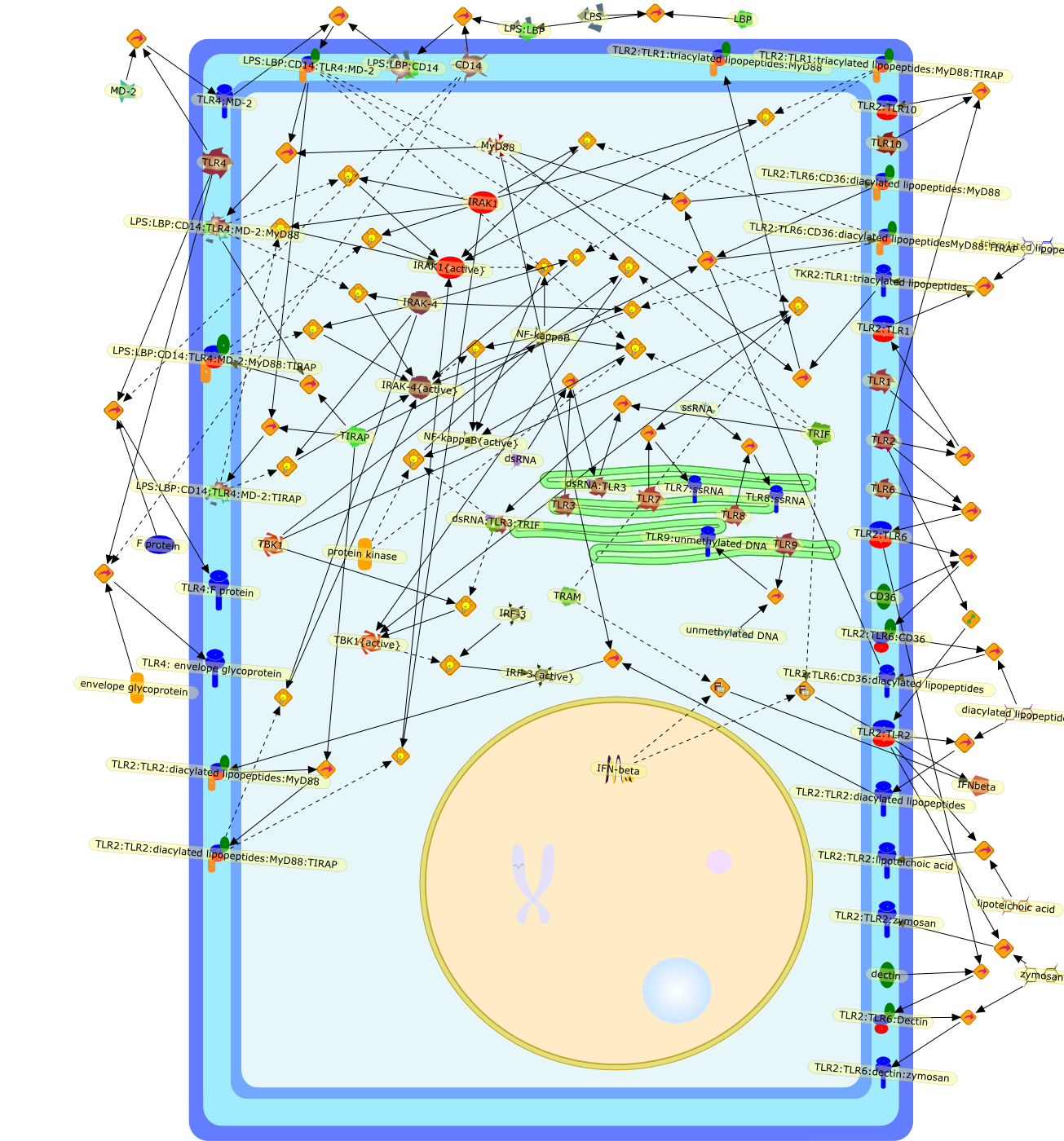
Many genes, and conceivably most genes, are constitutively expressed yet haveconditional functions. Their products are utilized only under specialcircumstances, and enforce homeostatic regulation. Mutations do not disclose thefunction of such genes unless the proper conditions are applied. The genes thatencode the Toll-like receptors (TLRs) fall into this category. The TLRsrepresent the principal sensors of infection in mammals. Absent infection,mammals have little need for the TLRs; they are essential only when microbesgain access to the interior milieu of the host. The function of the TLRs inmammals was first disclosed by a spontaneous mutation in a locus called Lps,when it was shown by positional cloning to be identical to Tlr4. Random germlinemutagenesis has since permitted an estimate of the total number of proteinsrequired for TLR signaling to the level of tumor necrosis factor (TNF) synthesisand activity, and has also shown that these sensors are extremely broad in theirability to detect microbes. Ultimately, the TLRs are responsible for mostinfection-related phenomena, both good and bad. These include the development offever, shock, and tissue injury, but also the activation of innate and adaptiveeffector mechanisms that lead to the elimination of microbes.
PMID
16001129
|