| Original Literature | Model OverView |
|---|---|
|
Publication
Title
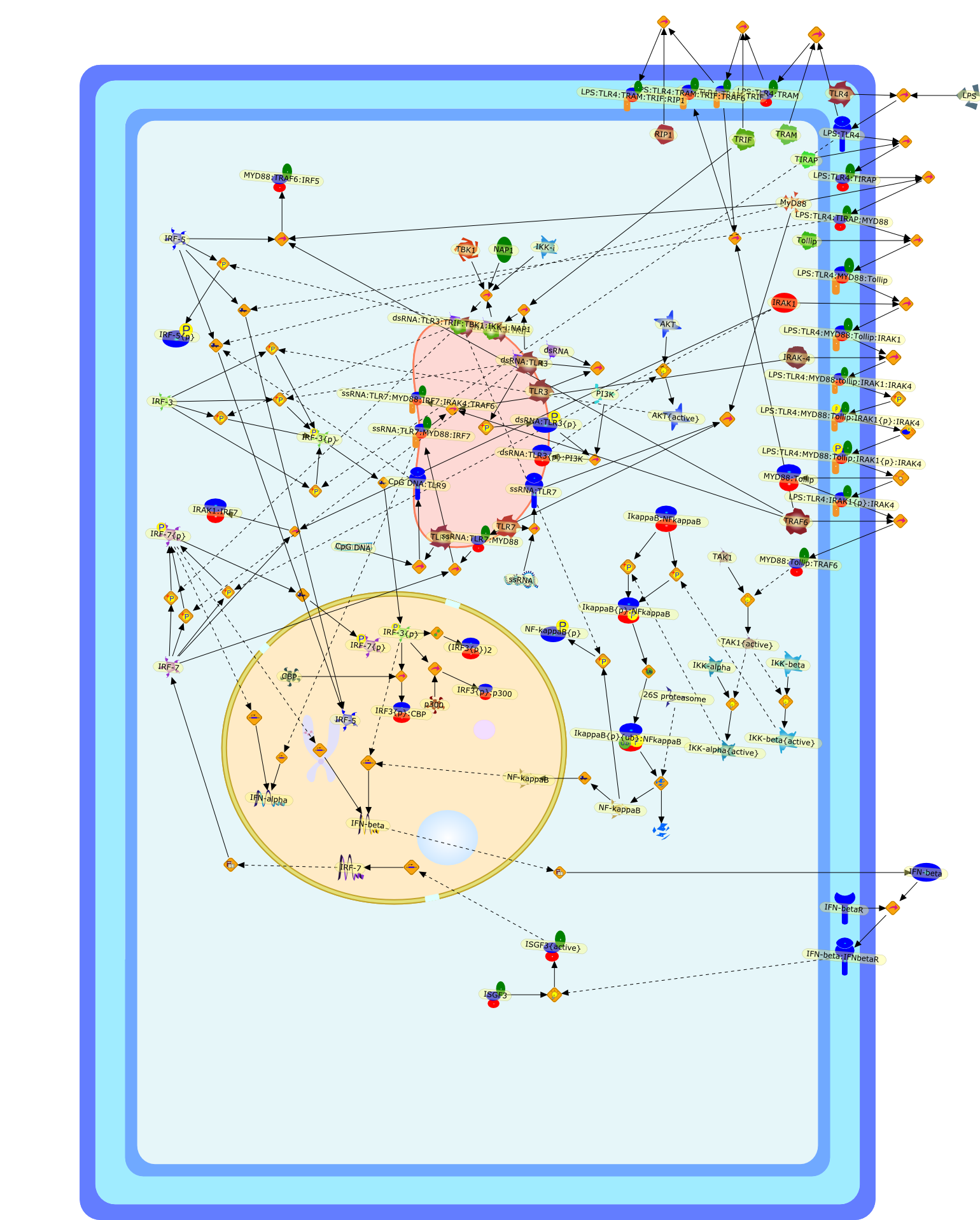
TLR signalling and activation of IRFs: revisiting old friends from the NF-kappaBpathway.
Affiliation
Department of Pharmacology, Conway Institute of Biomolecular and BiomedicalResearch, University College Dublin, Belfield, Dublin 4, Ireland.P.Moynagh@ucd.ie
Abstract
Toll-like receptors (TLRs) are crucially important in the sensing of infectiousagents. They serve to recognize pathogen-associated molecules and respond bytriggering the induction of specific profiles of proteins that are tailored tothe successful removal of the invading pathogens. The induction ofTLR-responsive genes is mediated by the activation of transcription factors, andmost interest has focussed on NF-kappaB, a transcription factor that isuniversally used by all TLRs. However, there has recently been a burgeoningeffort to increase our appreciation of the importance of members of theinterferon-regulatory factor (IRF) family in TLR signalling. This review willdiscuss the most recent findings relating to the regulation of IRF activity byTLRs and will highlight the rapidly increasing complexity of TLR signallingpathways.
PMID
16006187
|