| Original Literature | Model OverView |
|---|---|
|
Publication
Title
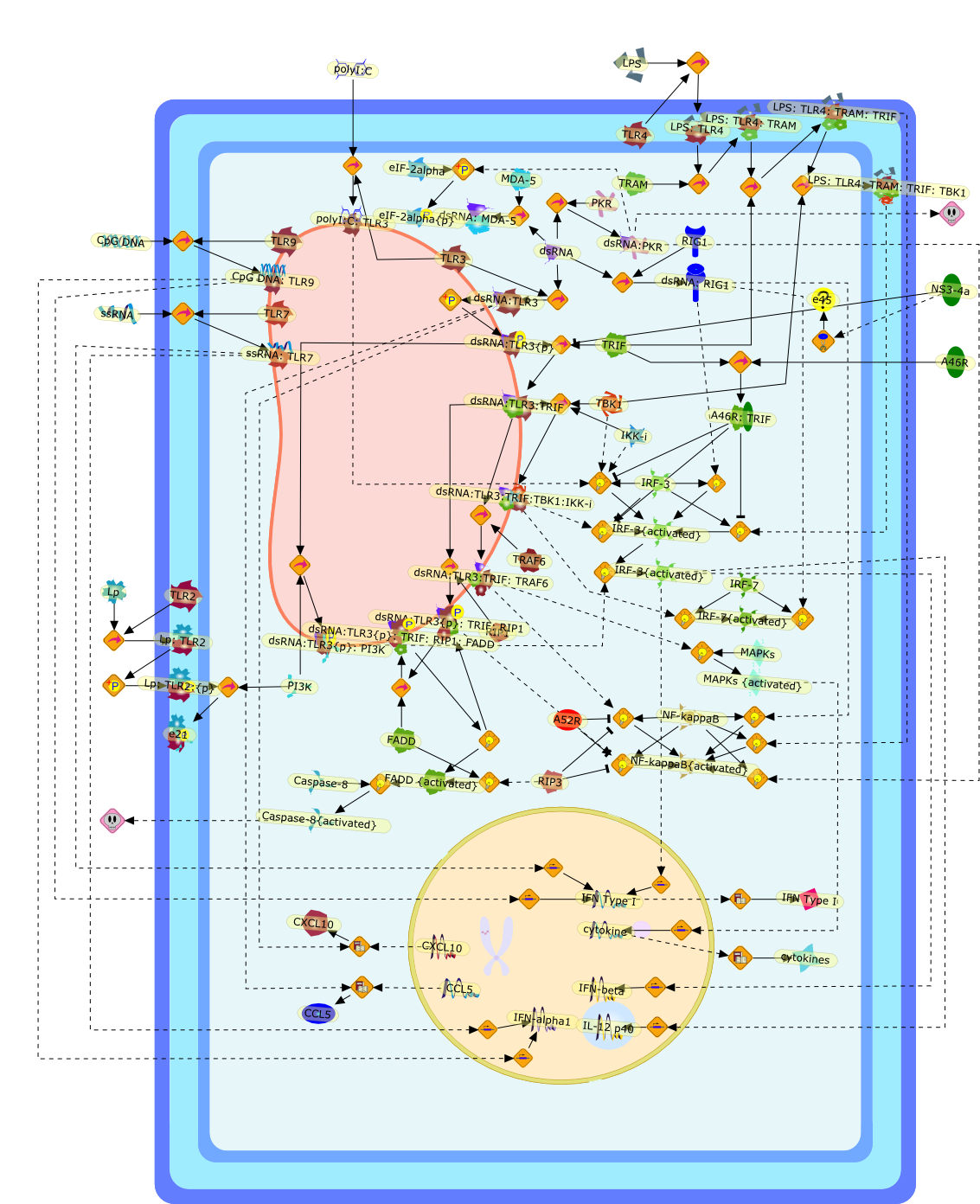
TLR3 in antiviral immunity: key player or bystander?
Affiliation
Viral Immune Evasion Group, Department of Biochemistry, Trinity College Dublin,College Green, Dublin 2, Ireland.
Abstract
Toll-like receptor 3 (TLR3), which recognizes double-stranded (ds)RNA, was thefirst identified antiviral TLR and, because dsRNA is a universal viral molecularpattern, TLR3 has been assumed to have a central role in the host response toviruses. However, this role has recently been questioned by in vivo studies andthe discovery of several other antiviral pattern-recognition receptors. In thisreview, the function of TLR3 in the context of these other receptors, namelyTLR7, 8 and 9 and the newly identified dsRNA-receptor retinoic-acid induciblegene-I (RIG-I) is discussed. Also, recent research concerning the expressionprofile of TLR3, its evasion by viruses and a potential role in crosspriming isaddressed, which reveals a clearer appreciation of the contribution of TLR3 toantiviral immunity.
PMID
16027039
|