| Original Literature | Model OverView |
|---|---|
|
Publication
Title
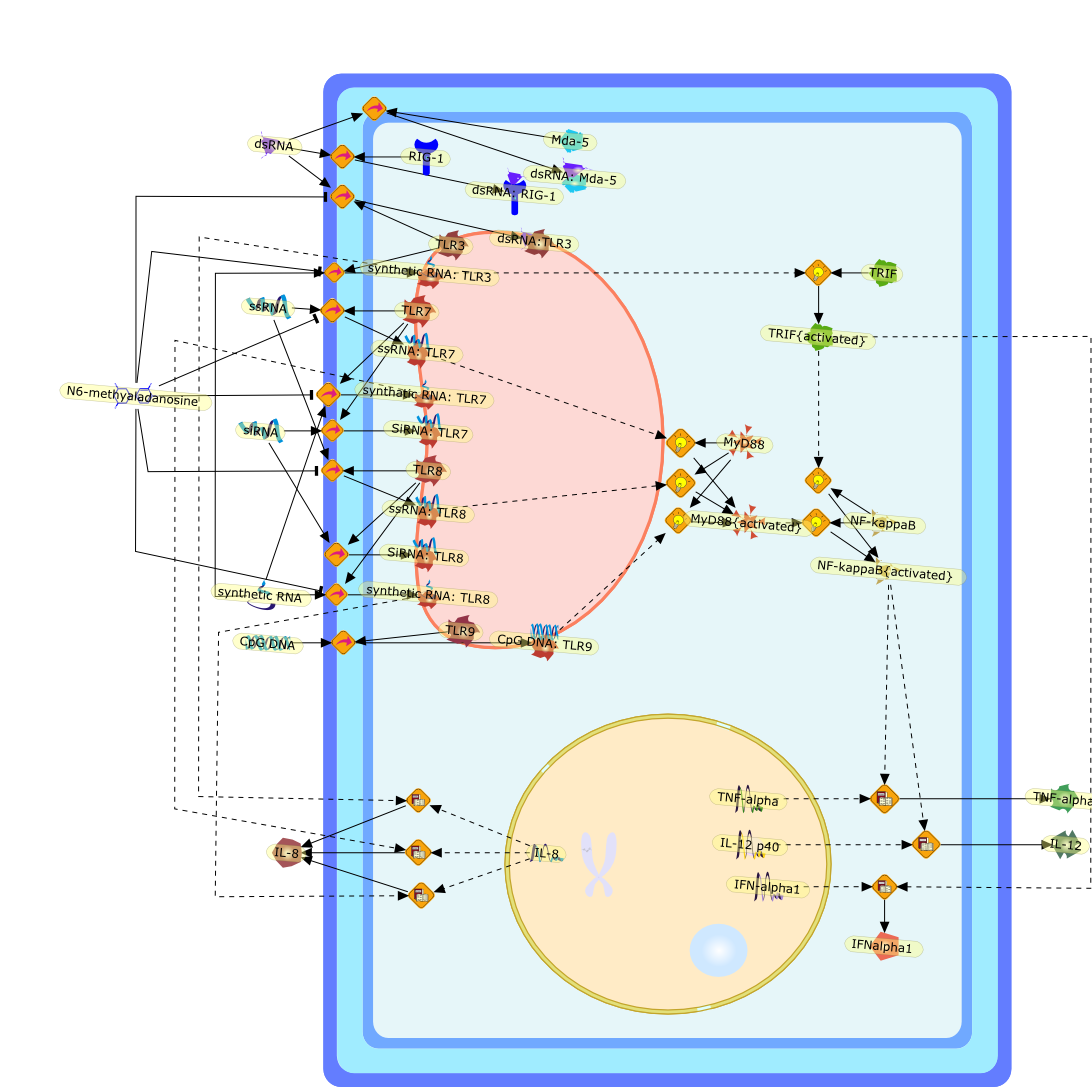
TLR ignores methylated RNA?
Affiliation
ERATO, Japan Science and Technology Agency (JST), Osaka University.
Abstract
CpG methylation of DNA silences TLR9-mediated innate immune recognition. In thisissue of Immunity, Kariko et al (2005) suggest that the innate immunerecognition of RNA by TLR3, TLR7, or TLR8 is in fact controlled by modificationof nucleotides, including methylation.
PMID
16111629
|