| Original Literature | Model OverView |
|---|---|
|
Publication
Title
ITAM-based signaling beyond the adaptive immune response.
Affiliation
Department of Computer Science, Corvinus University, Budapest, Hungary.
Abstract
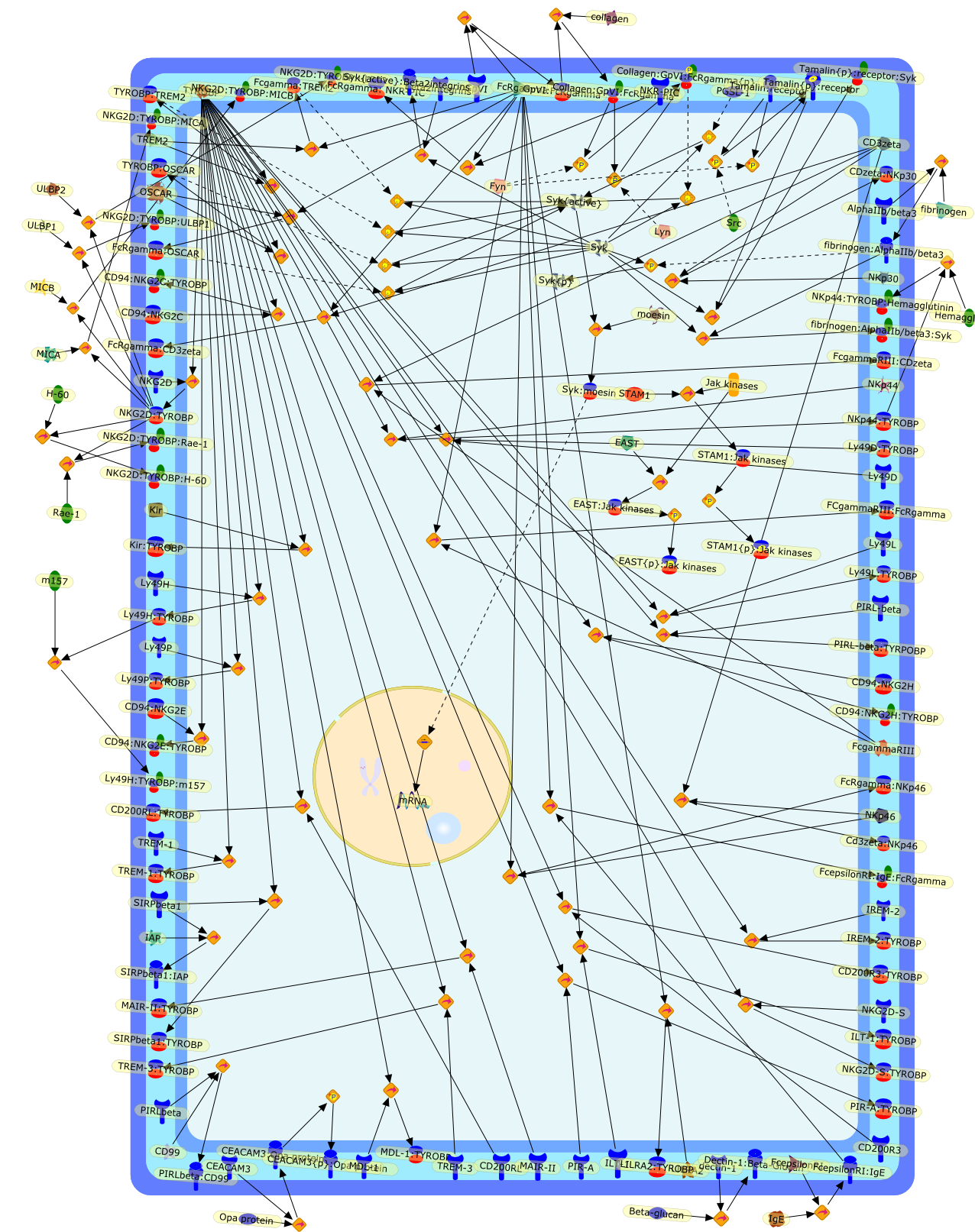
Classical immunoreceptors like lymphocyte antigen receptors and Fc-receptors(FcR) are central players of the adaptive immune response. These receptorsutilize a common signal transduction mechanism, which relies on immunoreceptortyrosine-based activation motifs (ITAMs) present in the receptor complex. Uponligand binding to the receptors, tyrosines within the ITAM sequence arephosphorylated by Src-family kinases, leading to an SH2-domain mediatedrecruitment and activation of the Syk or the related ZAP-70 tyrosine kinase.These kinases then initiate further downstream signaling events. Here we reviewrecent evidence indicating that components of this ITAM-based signalingmachinery are also present in a number of non-lymphoid or even non-immune celltypes and they participate in diverse biological functions beyond the adaptiveimmune response, including innate immune mechanisms, platelet activation, boneresorption or tumor development. These results suggest that the ITAM-basedsignaling paradigm has much wider implications than previously anticipated.
PMID
16332394
|