| Original Literature | Model OverView |
|---|---|
|
Publication
Title
Lipopolysaccharide signaling in endothelial cells.
Affiliation
Department of Medical Biophysics, British Columbia Cancer Agency, Vancouver,British Columbia, Canada.
Abstract
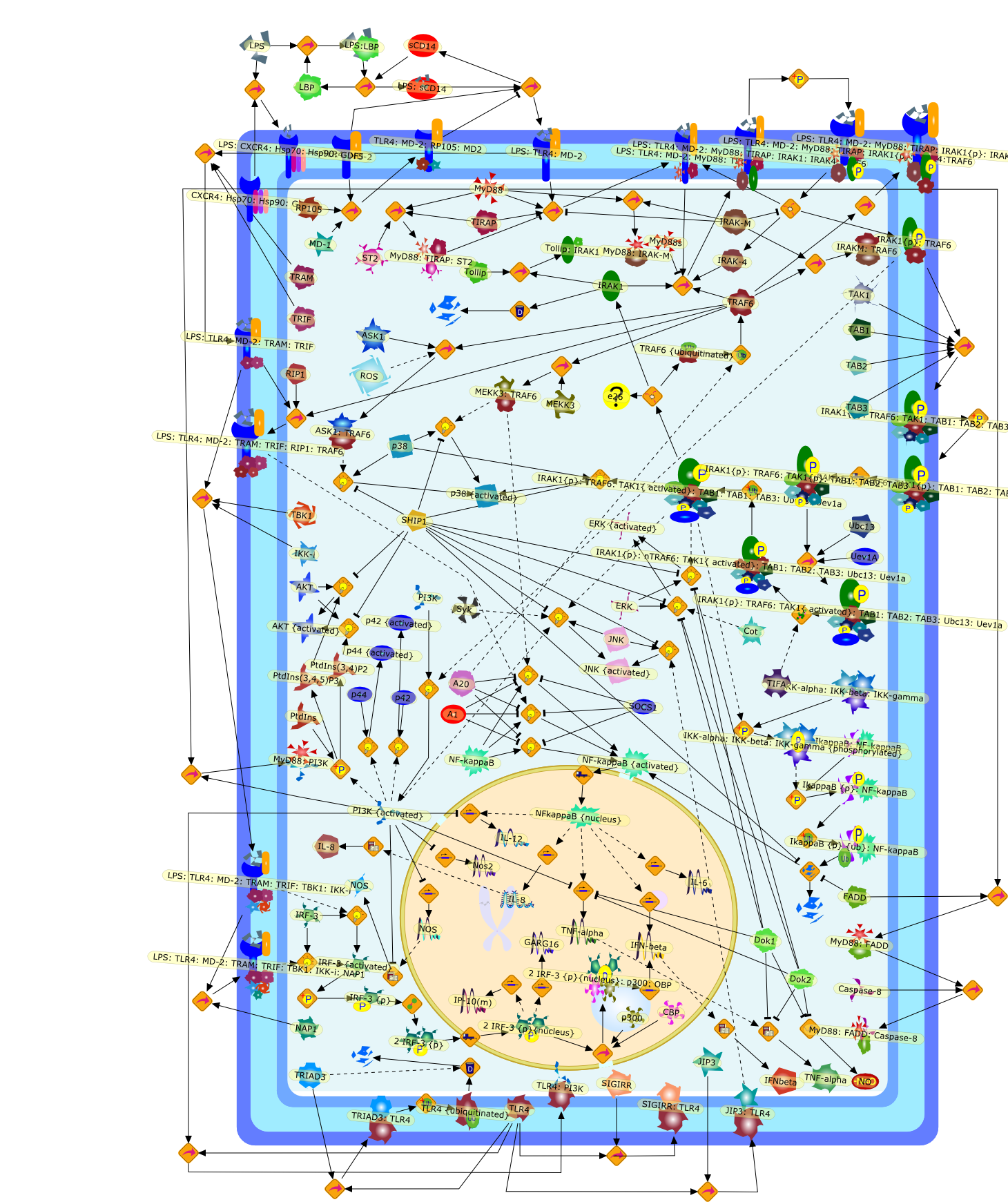
Sepsis is the systemic immune response to severe bacterial infection. The innateimmune recognition of bacterial and viral products is mediated by a family oftransmembrane receptors known as Toll-like receptors (TLRs). In endothelialcells, exposure to lipopolysaccharide (LPS), a major cell wall constituent ofGram-negative bacteria, results in endothelial activation through a receptorcomplex consisting of TLR4, CD14 and MD2. Recruitment of the adaptor proteinmyeloid differentiation factor (MyD88) initiates an MyD88-dependent pathway thatculminates in the early activation of nuclear factor-kappaB (NF-kappaB) and themitogen-activated protein kinases. In parallel, a MyD88-independent pathwayresults in a late-phase activation of NF-kappaB. The outcome is the productionof various proinflammatory mediators and ultimately cellular injury, leading tothe various vascular sequelae of sepsis. This review will focus on the signalingpathways initiated by LPS binding to the TLR4 receptor in endothelial cells andthe coordinated regulation of this pathway.
PMID
16357866
|