| Original Literature | Model OverView |
|---|---|
|
Publication
Title
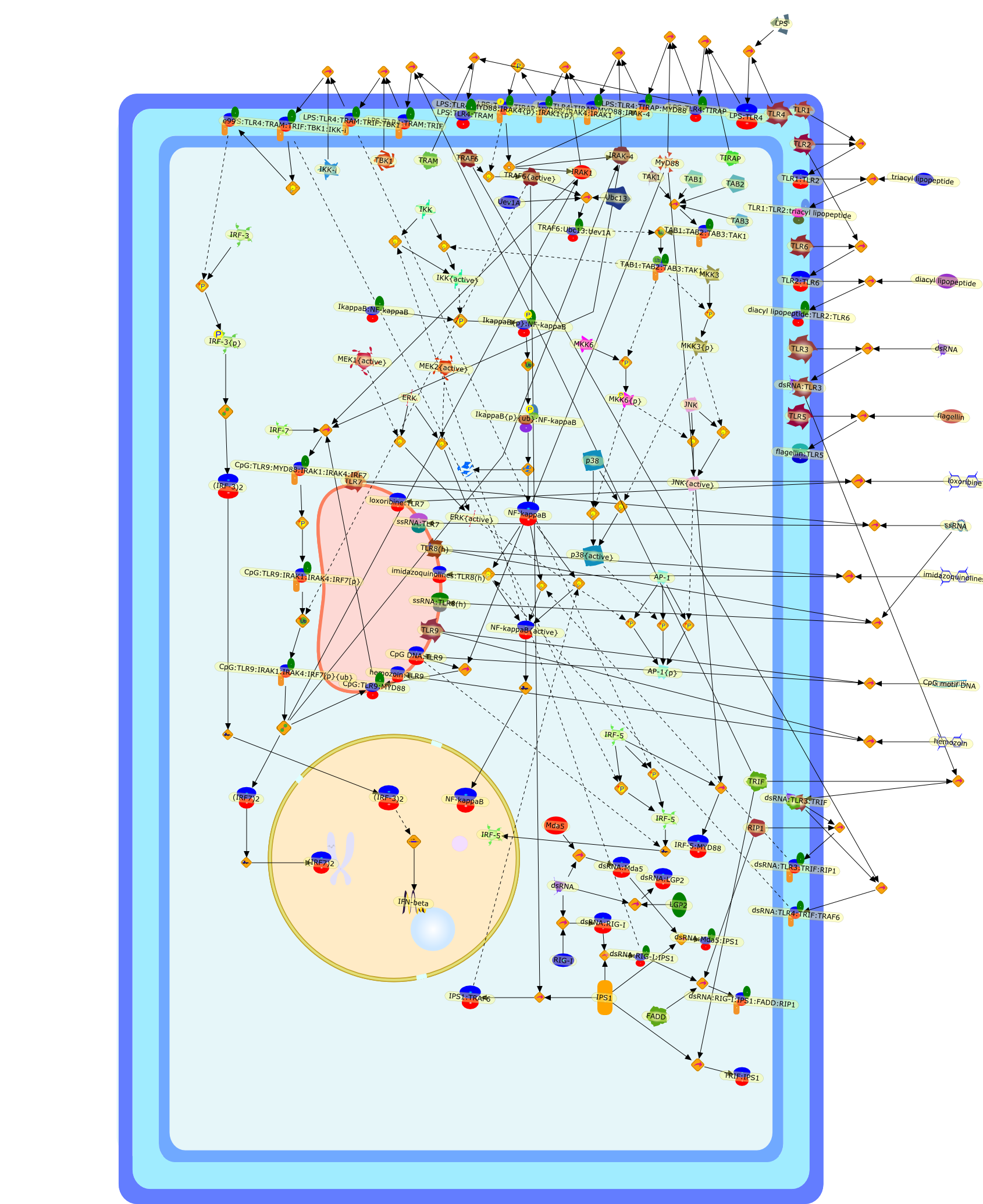
TLR signaling.
Affiliation
Exploratory Research for Advanced Technology, Japan Science and TechnologyAgency, 3-1 Yamada-oka, Suita, Osaka 565-0871, Japan.
Abstract
The Toll-like receptor (TLR) family plays an instructive role in innate immuneresponses against microbial pathogens, as well as the subsequent induction ofadaptive immune responses. TLRs recognize specific molecular patterns found in abroad range of microbial pathogens such as bacteria and viruses, triggeringinflammatory and antiviral responses and dendritic cell maturation, which resultin the eradication of invading pathogens. Individual TLRs interact withdifferent combinations of adapter proteins and activate various transcriptionfactors such as nuclear factor (NF)-kappaB, activating protein-1 and interferonregulatory factors, driving a specific immune response. This review outlines therecent advances in our understanding of TLR-signaling pathways and their rolesin immune responses. Further, we also discuss a new concept of TLR-independentmechanisms for recognition of microbial pathogens.
PMID
16410796
|